
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 11,469,362 – 11,469,461 |
| Length | 99 |
| Max. P | 0.578803 |

| Location | 11,469,362 – 11,469,461 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 82.59 |
| Mean single sequence MFE | -27.99 |
| Consensus MFE | -18.67 |
| Energy contribution | -21.20 |
| Covariance contribution | 2.53 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.578803 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11469362 99 - 27905053 AAUAAAGGAGGAACCA---CUGACAUUGCUACCGAUCGAUCG----UCUUGGGUAAUCCCAUCCAGAGCUCA--GCUGAGAUCGGUGAGCAAUGGACUGCAGAUAAGA- ......((.....)).---(((.((((((((((((((.....----...((((....))))..(((......--.))).))))))).))))))).....)))......- ( -30.90) >DroSec_CAF1 3528 99 - 1 AACAAAGGAGGAACCA---CUGACAUUGCUACCGAUCGAUCG----UCUUGGGUAAUCCCAUCCAGAGCUCA--GCUGAGAUCGGUGAGCAAUGGACUGCAGAUAAGA- ......((.....)).---(((.((((((((((((((.....----...((((....))))..(((......--.))).))))))).))))))).....)))......- ( -30.90) >DroSim_CAF1 3742 99 - 1 AACAAAGGAGGAACCA---CUGACAUUGCUACCGAUCGAUCG----UCUUGGGUAAUCCCAUCCAGAGCUCA--GCUGAGAUCGGUGAGCAAUGGACUGCAGAUAAAA- ......((.....)).---(((.((((((((((((((.....----...((((....))))..(((......--.))).))))))).))))))).....)))......- ( -30.90) >DroEre_CAF1 5543 91 - 1 AAGAAAAG------------UGACAUUGCUACCGAUCGAUCG----CCUUGGGUAAUCCCAUCCAGAGCCCA--GCUGAGAUCGGUGAGCAAUGCAUUGCAGAUAAGAG ..(...((------------((.((((((((((((((....(----(..(((((.............)))))--))...))))))).))))))))))).)......... ( -30.82) >DroYak_CAF1 853 102 - 1 AACAAAGGAGGAACCA---CUGGCAUGGCUACCGAUCGAUCG----CCUUGGGUAAUCCCAUCCAGAGCUCAGAGCUGAGAUCGCUGAGCAAUGGAUUGCAGAUAAGAG ......(((...(((.---..(((..((...))((....)))----))...)))..))).(((((..((((((...........))))))..)))))............ ( -26.70) >DroAna_CAF1 92374 90 - 1 AACAAAGGAGUAACCACUACUGACACUGCUACCGAUCGAUCGAUCCUCUUGGGUAAUCCAAUCCAAAACUCA--GCUGAGCU----------------CCAGAUAAGA- ......(((((...((...((((..........((((....))))...((((((......))))))...)))--).)).)))----------------))........- ( -17.70) >consensus AACAAAGGAGGAACCA___CUGACAUUGCUACCGAUCGAUCG____UCUUGGGUAAUCCCAUCCAGAGCUCA__GCUGAGAUCGGUGAGCAAUGGACUGCAGAUAAGA_ ...................(((.((((((((((((((...........((((((......))))))(((.....)))..))))))).))))))).....)))....... (-18.67 = -21.20 + 2.53)
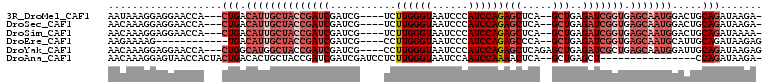

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:27:06 2006