| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 1,634,698 – 1,634,796 |
| Length | 98 |
| Max. P | 0.854766 |

| Location | 1,634,698 – 1,634,796 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 77.23 |
| Mean single sequence MFE | -32.95 |
| Consensus MFE | -18.72 |
| Energy contribution | -19.12 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.80 |
| SVM RNA-class probability | 0.854766 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
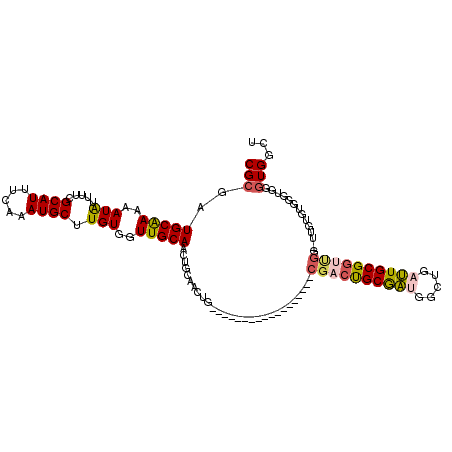
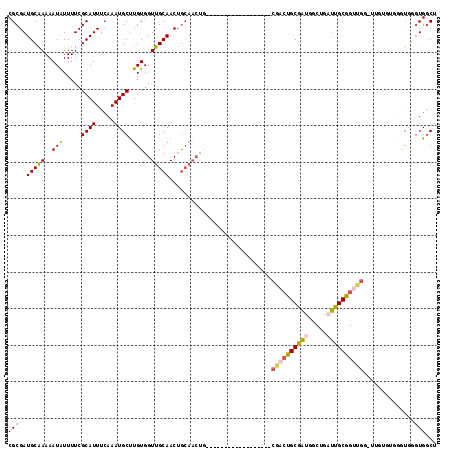
>3R_DroMel_CAF1 1634698 98 - 27905053 CGCGAUGCAAAAAUAUUUUCGCAUUUCAAAUGCUUGUGGUUGCAACUGCAACUG------------------CGACUGCGAUGGCUGAUUGCGGUUGG-UUGUGUGGGUGGGUGGCU .((.((.((...((((..((((((.....))))..(..(((((....)))))..------------------)(((((((((.....)))))))))))-..))))...)).)).)). ( -33.90) >DroSec_CAF1 14248 94 - 1 CGCGAUGCAAAAAUAUUUUCGCAUUUCAAAUGCUUGUGGUUGCAACUGCAACUG------------------CGACUGCGAUGGCUGAUUGCGGUUGG-UUGUGUGG----GUGGCU .((.((.((..(((......((((.....))))..(..(((((....)))))..------------------)(((((((((.....))))))))).)-))...)).----)).)). ( -30.30) >DroSim_CAF1 14172 98 - 1 CGCGAUGCAAAAAUAUUUUCGCAUUUCAAAUGCUUGUGGUUGCAACUGCAACUG------------------CGACUGCGAUGGCUGAUUGCGGUUGG-UUGUGUGGGUGGGUGGCU .((.((.((...((((..((((((.....))))..(..(((((....)))))..------------------)(((((((((.....)))))))))))-..))))...)).)).)). ( -33.90) >DroEre_CAF1 15334 84 - 1 CGCGAUGCAAAAAUAUUUUCGCAUUUCAAAUGCCUGUGGUUGCAACUGCAAC------------------------UGCGAUGGCUGAUUGCGGUUG---------GGUGGCUGGCU .((.((.(((........(((((..(((...(((((..(((((....)))))------------------------..))..)))))).))))))))---------.)).))..... ( -27.10) >DroYak_CAF1 12971 108 - 1 CGCGAUGCAAAAAUGUUUUCGCAUUUGAAAUGCUUGUGGUUGCAACUGCAACUGCAACUGCAAAUGCGAUUGCGACUGCGACGGCUGAUUGCGGUUG---------UGUGGGUGGCU .((.((.((....(((..(((((((((....(.(((..(((((....)))))..))))..)))))))))..)))...(((((.((.....)).))))---------).)).)).)). ( -43.70) >DroAna_CAF1 14312 93 - 1 CGCGAUGCGAAAAUAUUUUCGCAUUUCAAAUGCUUGUGGUUGCAACAA------------------------CGCCCGCAUGGGCUGGGUGCGGUCGAGAGGUCUACAUAUGUGUCU .(.(((((((((....))))))))).)..((((.((((...(((.(..------------------------.((((....))))..).)))(..(....)..)..)))).)))).. ( -28.80) >consensus CGCGAUGCAAAAAUAUUUUCGCAUUUCAAAUGCUUGUGGUUGCAACUGCAACUG__________________CGACUGCGAUGGCUGAUUGCGGUUGG_UUGUGUGGGUGGGUGGCU (((..(((((..(((.....((((.....)))).)))..)))))............................((((((((((.....))))))))))..............)))... (-18.72 = -19.12 + 0.39)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:43:07 2006