
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 11,464,742 – 11,464,938 |
| Length | 196 |
| Max. P | 0.983835 |

| Location | 11,464,742 – 11,464,859 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.33 |
| Mean single sequence MFE | -28.12 |
| Consensus MFE | -24.23 |
| Energy contribution | -24.48 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.716904 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11464742 117 + 27905053 AUAUAUAUUACAUUAU---CCCUAGUCUGAUUUCUGGCAGUGCAUUUAUACACUUGUGGCAACGUAUUUCCGACAGGAAACCAUUCGCAGCCACCGCCAAAAUGUGGCAACAUCGCCAGA ................---.............((((((((((........)))).(((((..((..(((((....))))).....))..))))).((((.....))))......)))))) ( -30.80) >DroSec_CAF1 198604 120 + 1 AUAUAUAUUACAUUAUUACUCCUAGGCUGAUUUCUUGCAGUGCAUUUAUACACUUGUGGCAACGUAUUUCCGACAGGAAACCAUUCGCAGCCACCGCCAAAAUGUGGCAACAUCGCCAGA ........................(((.(((.......((((........)))).(((((..((..(((((....))))).....))..))))).((((.....))))...))))))... ( -26.40) >DroSim_CAF1 193752 120 + 1 AUAUAUAUUACAUUAUUAUUGCUUGGCUGAUUUUUUGCAGUGCAUUUAUACACUUGUGGCAACGUAUUUCCGACAGGAAACCAUUCGCAGCCACCGCCAAAAUGUGGCAACAUCGCCAGA ......................(((((...........((((........)))).(((((..((..(((((....))))).....))..))))).)))))....((((......)))).. ( -27.70) >DroYak_CAF1 190907 113 + 1 ----AUAUUACCUAAU---UCCUAGGCUGAUUUCUUGCAGUGCAUUUACACACUUGUGGCAACGUAUUUCCGACAGGAAACCAUUCGCAGCCACCGCCAAAAUGUGGCAACAACGCCAGA ----..((((((((..---...)))).)))).(((.((.(((......)))....(((((..((..(((((....))))).....))..))))).((((.....))))......)).))) ( -27.60) >consensus AUAUAUAUUACAUUAU___UCCUAGGCUGAUUUCUUGCAGUGCAUUUAUACACUUGUGGCAACGUAUUUCCGACAGGAAACCAUUCGCAGCCACCGCCAAAAUGUGGCAACAUCGCCAGA ........................(((...........((((........)))).(((((..((..(((((....))))).....))..))))).)))......((((......)))).. (-24.23 = -24.48 + 0.25)



| Location | 11,464,742 – 11,464,859 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.33 |
| Mean single sequence MFE | -36.65 |
| Consensus MFE | -31.86 |
| Energy contribution | -31.92 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.58 |
| SVM RNA-class probability | 0.965470 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11464742 117 - 27905053 UCUGGCGAUGUUGCCACAUUUUGGCGGUGGCUGCGAAUGGUUUCCUGUCGGAAAUACGUUGCCACAAGUGUAUAAAUGCACUGCCAGAAAUCAGACUAGGG---AUAAUGUAAUAUAUAU ((((((.....(((((.....)))))(((((.(((....((((((....)))))).))).))))).((((((....)))))))))))).............---................ ( -39.00) >DroSec_CAF1 198604 120 - 1 UCUGGCGAUGUUGCCACAUUUUGGCGGUGGCUGCGAAUGGUUUCCUGUCGGAAAUACGUUGCCACAAGUGUAUAAAUGCACUGCAAGAAAUCAGCCUAGGAGUAAUAAUGUAAUAUAUAU (((((((((..(((((.....)))))(((((.(((....((((((....)))))).))).))))).((((((....)))))).......))).))).))).................... ( -35.20) >DroSim_CAF1 193752 120 - 1 UCUGGCGAUGUUGCCACAUUUUGGCGGUGGCUGCGAAUGGUUUCCUGUCGGAAAUACGUUGCCACAAGUGUAUAAAUGCACUGCAAAAAAUCAGCCAAGCAAUAAUAAUGUAAUAUAUAU ..(((((....)))))...((((((.(((((.(((....((((((....)))))).))).))))).((((((....))))))...........))))))..................... ( -36.20) >DroYak_CAF1 190907 113 - 1 UCUGGCGUUGUUGCCACAUUUUGGCGGUGGCUGCGAAUGGUUUCCUGUCGGAAAUACGUUGCCACAAGUGUGUAAAUGCACUGCAAGAAAUCAGCCUAGGA---AUUAGGUAAUAU---- (((.((.....(((((.....)))))(((((.(((....((((((....)))))).))).))))).(((((......))))))).))).....((((((..---.)))))).....---- ( -36.20) >consensus UCUGGCGAUGUUGCCACAUUUUGGCGGUGGCUGCGAAUGGUUUCCUGUCGGAAAUACGUUGCCACAAGUGUAUAAAUGCACUGCAAGAAAUCAGCCUAGGA___AUAAUGUAAUAUAUAU ..(((((....)))))......(((.(((((.(((....((((((....)))))).))).))))).((((((....))))))...........)))........................ (-31.86 = -31.92 + 0.06)

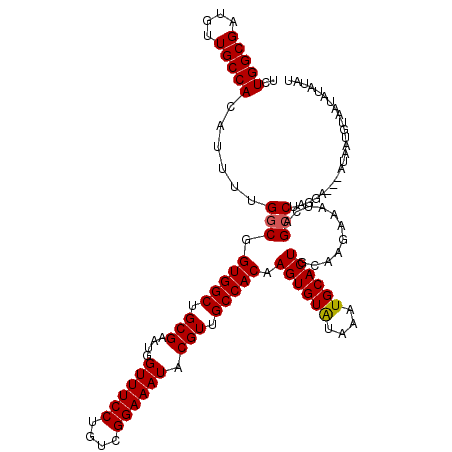
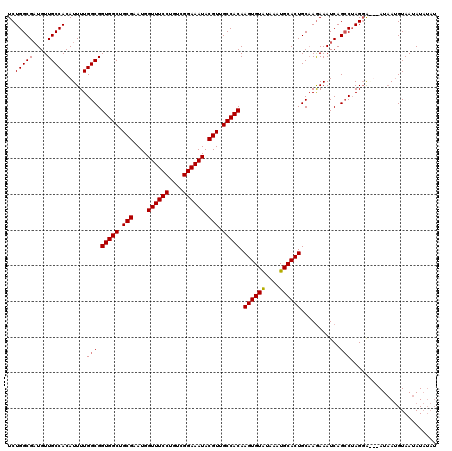
| Location | 11,464,779 – 11,464,899 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.50 |
| Mean single sequence MFE | -35.60 |
| Consensus MFE | -35.51 |
| Energy contribution | -35.58 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.86 |
| Structure conservation index | 1.00 |
| SVM decision value | 1.96 |
| SVM RNA-class probability | 0.983835 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11464779 120 + 27905053 UGCAUUUAUACACUUGUGGCAACGUAUUUCCGACAGGAAACCAUUCGCAGCCACCGCCAAAAUGUGGCAACAUCGCCAGAGACAACAGAGGAUGAAGGAAGCUUACUUCCUAUUCCUCUA .((............(((((..((..(((((....))))).....))..))))).((((.....))))......))..........((((((...((((((....))))))..)))))). ( -36.70) >DroSec_CAF1 198644 120 + 1 UGCAUUUAUACACUUGUGGCAACGUAUUUCCGACAGGAAACCAUUCGCAGCCACCGCCAAAAUGUGGCAACAUCGCCAGAGACAACAGAGGAUGAAGGAAGCUUACUUCCUAUUCCUCUA .((............(((((..((..(((((....))))).....))..))))).((((.....))))......))..........((((((...((((((....))))))..)))))). ( -36.70) >DroSim_CAF1 193792 120 + 1 UGCAUUUAUACACUUGUGGCAACGUAUUUCCGACAGGAAACCAUUCGCAGCCACCGCCAAAAUGUGGCAACAUCGCCAGAGACAACAGAGGAUGAAGGAAGCUUACUUCCUAUUCCUCUA .((............(((((..((..(((((....))))).....))..))))).((((.....))))......))..........((((((...((((((....))))))..)))))). ( -36.70) >DroYak_CAF1 190940 120 + 1 UGCAUUUACACACUUGUGGCAACGUAUUUCCGACAGGAAACCAUUCGCAGCCACCGCCAAAAUGUGGCAACAACGCCAGAGACAACGGAGGAUAAAGGAAGCUGACUUCCUAUUCCUAUA .((............(((((..((..(((((....))))).....))..))))).((((.....))))......))............((((...((((((....))))))..))))... ( -32.30) >consensus UGCAUUUAUACACUUGUGGCAACGUAUUUCCGACAGGAAACCAUUCGCAGCCACCGCCAAAAUGUGGCAACAUCGCCAGAGACAACAGAGGAUGAAGGAAGCUUACUUCCUAUUCCUCUA .((............(((((..((..(((((....))))).....))..))))).((((.....))))......))..........((((((...((((((....))))))..)))))). (-35.51 = -35.58 + 0.06)


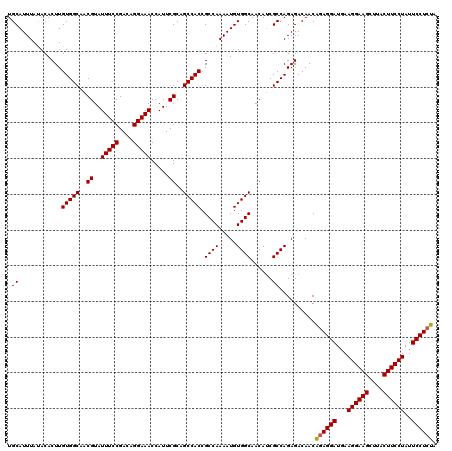
| Location | 11,464,779 – 11,464,899 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.50 |
| Mean single sequence MFE | -43.25 |
| Consensus MFE | -41.97 |
| Energy contribution | -42.47 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.94 |
| Structure conservation index | 0.97 |
| SVM decision value | 1.83 |
| SVM RNA-class probability | 0.979237 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11464779 120 - 27905053 UAGAGGAAUAGGAAGUAAGCUUCCUUCAUCCUCUGUUGUCUCUGGCGAUGUUGCCACAUUUUGGCGGUGGCUGCGAAUGGUUUCCUGUCGGAAAUACGUUGCCACAAGUGUAUAAAUGCA .((((((..((((((....))))))...))))))(((((.....)))))..(((((.....)))))(((((.(((....((((((....)))))).))).)))))..((((....)))). ( -44.20) >DroSec_CAF1 198644 120 - 1 UAGAGGAAUAGGAAGUAAGCUUCCUUCAUCCUCUGUUGUCUCUGGCGAUGUUGCCACAUUUUGGCGGUGGCUGCGAAUGGUUUCCUGUCGGAAAUACGUUGCCACAAGUGUAUAAAUGCA .((((((..((((((....))))))...))))))(((((.....)))))..(((((.....)))))(((((.(((....((((((....)))))).))).)))))..((((....)))). ( -44.20) >DroSim_CAF1 193792 120 - 1 UAGAGGAAUAGGAAGUAAGCUUCCUUCAUCCUCUGUUGUCUCUGGCGAUGUUGCCACAUUUUGGCGGUGGCUGCGAAUGGUUUCCUGUCGGAAAUACGUUGCCACAAGUGUAUAAAUGCA .((((((..((((((....))))))...))))))(((((.....)))))..(((((.....)))))(((((.(((....((((((....)))))).))).)))))..((((....)))). ( -44.20) >DroYak_CAF1 190940 120 - 1 UAUAGGAAUAGGAAGUCAGCUUCCUUUAUCCUCCGUUGUCUCUGGCGUUGUUGCCACAUUUUGGCGGUGGCUGCGAAUGGUUUCCUGUCGGAAAUACGUUGCCACAAGUGUGUAAAUGCA ...((((..((((((....))))))...)))).(((((....(((((....))))).....)))))(((((.(((....((((((....)))))).))).)))))..((((....)))). ( -40.40) >consensus UAGAGGAAUAGGAAGUAAGCUUCCUUCAUCCUCUGUUGUCUCUGGCGAUGUUGCCACAUUUUGGCGGUGGCUGCGAAUGGUUUCCUGUCGGAAAUACGUUGCCACAAGUGUAUAAAUGCA (((((((..((((((....))))))...))))))).........(((....(((((.....)))))(((((.(((....((((((....)))))).))).)))))...........))). (-41.97 = -42.47 + 0.50)

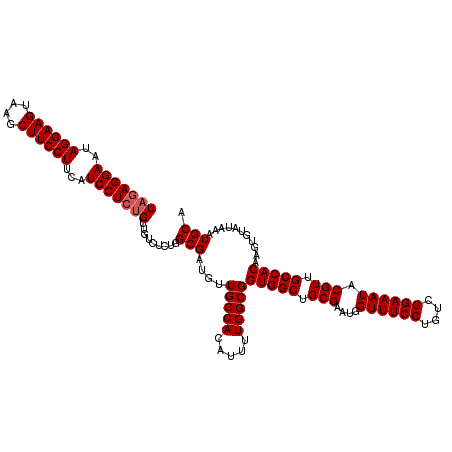

| Location | 11,464,819 – 11,464,938 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.22 |
| Mean single sequence MFE | -30.77 |
| Consensus MFE | -25.60 |
| Energy contribution | -26.13 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.21 |
| SVM RNA-class probability | 0.930880 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11464819 119 + 27905053 CCAUUCGCAGCCACCGCCAAAAUGUGGCAACAUCGCCAGAGACAACAGAGGAUGAAGGAAGCUUACUUCCUAUUCCUCUAUGUAGACCCGCGCCUGGAACUCCACAUACACAUCCUCGC- (((..(((.(((((.........))))).............(((..((((((...((((((....))))))..)))))).)))......)))..)))......................- ( -32.20) >DroSec_CAF1 198684 120 + 1 CCAUUCGCAGCCACCGCCAAAAUGUGGCAACAUCGCCAGAGACAACAGAGGAUGAAGGAAGCUUACUUCCUAUUCCUCUAUGUAGACCCGCGCCUGGAACUCCUCAUACUCCUCCUCGCA (((..(((.(((((.........))))).............(((..((((((...((((((....))))))..)))))).)))......)))..)))....................... ( -32.20) >DroYak_CAF1 190980 101 + 1 CCAUUCGCAGCCACCGCCAAAAUGUGGCAACAACGCCAGAGACAACGGAGGAUAAAGGAAGCUGACUUCCUAUUCCUAUAUGG-------------------UAUGUACUCCUCCUCGCA ......((.(((((.........)))))..................((((((...((((((....))))))...((.....))-------------------.......))))))..)). ( -27.90) >consensus CCAUUCGCAGCCACCGCCAAAAUGUGGCAACAUCGCCAGAGACAACAGAGGAUGAAGGAAGCUUACUUCCUAUUCCUCUAUGUAGACCCGCGCCUGGAACUCCACAUACUCCUCCUCGCA ......((.(((((.........)))))..........(((.....((((((...((((((....))))))..))))))................((....))...........))))). (-25.60 = -26.13 + 0.53)


| Location | 11,464,819 – 11,464,938 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.22 |
| Mean single sequence MFE | -39.87 |
| Consensus MFE | -31.17 |
| Energy contribution | -32.17 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.78 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11464819 119 - 27905053 -GCGAGGAUGUGUAUGUGGAGUUCCAGGCGCGGGUCUACAUAGAGGAAUAGGAAGUAAGCUUCCUUCAUCCUCUGUUGUCUCUGGCGAUGUUGCCACAUUUUGGCGGUGGCUGCGAAUGG -((.(((((.((..(.(((....))).)..)).))).((((((((((..((((((....))))))...))))))).)))..)).))..(((.(((((.........))))).)))..... ( -44.20) >DroSec_CAF1 198684 120 - 1 UGCGAGGAGGAGUAUGAGGAGUUCCAGGCGCGGGUCUACAUAGAGGAAUAGGAAGUAAGCUUCCUUCAUCCUCUGUUGUCUCUGGCGAUGUUGCCACAUUUUGGCGGUGGCUGCGAAUGG (((........))).........(((..(((((..((((((((((((..((((((....))))))...))))))))((((..(((((....)))))......)))))))))))))..))) ( -40.60) >DroYak_CAF1 190980 101 - 1 UGCGAGGAGGAGUACAUA-------------------CCAUAUAGGAAUAGGAAGUCAGCUUCCUUUAUCCUCCGUUGUCUCUGGCGUUGUUGCCACAUUUUGGCGGUGGCUGCGAAUGG .((((((((((.......-------------------((.....))...((((((....))))))...)))))).)))).....((((..((((((.....))))))..)).))...... ( -34.80) >consensus UGCGAGGAGGAGUAUGUGGAGUUCCAGGCGCGGGUCUACAUAGAGGAAUAGGAAGUAAGCUUCCUUCAUCCUCUGUUGUCUCUGGCGAUGUUGCCACAUUUUGGCGGUGGCUGCGAAUGG .((.((((.................((((....))))..((((((((..((((((....))))))...))))))))..)).)).))..(((.(((((.........))))).)))..... (-31.17 = -32.17 + 1.00)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:26:58 2006