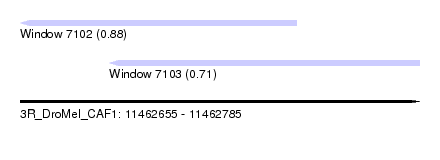
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 11,462,655 – 11,462,785 |
| Length | 130 |
| Max. P | 0.883903 |

| Location | 11,462,655 – 11,462,745 |
|---|---|
| Length | 90 |
| Sequences | 3 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 77.06 |
| Mean single sequence MFE | -17.37 |
| Consensus MFE | -14.13 |
| Energy contribution | -14.47 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.93 |
| SVM RNA-class probability | 0.883903 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11462655 90 - 27905053 UAAAACUCAGUUGAUUCGGAUUUCUCAGAAAUCCUUCUGAAGAAAUCUGAGUGCUUUU--------CAA-------UUUAU----AUUCUGUUUUGCCAAAUACGUAGA ((((((..((((((((((((((((((((((....))))).)))))))))))......)--------)))-------))...----.....))))))............. ( -19.70) >DroSec_CAF1 196455 109 - 1 UAAAACACAGUUGAUUCGGAUUUCUCAGAAAUCCUUCUGAAGAAAUCAGAGUGCUCUUUAACCCUUCAAUCUACAAUUCAUCUCUCUUCUGUUAUUCCAAAUACGUUCG ......((((..((....((((((((((((....))))).)))))))((((....)))).........................))..))))................. ( -16.60) >DroSim_CAF1 191609 109 - 1 AAAAACUCAGUUUAUUCGAAUUUCUCAGAAAUUCUUCUGAAGAAAUCAGAGUGCUCUUUAACCCUUCAAUUUACAAUUUAUCGCAAUUCUGUAAUGCAAAAUAUGUUCG ........((..(((((..(((((((((((....))))).))))))..)))))..)).........................(((.........)))............ ( -15.80) >consensus UAAAACUCAGUUGAUUCGGAUUUCUCAGAAAUCCUUCUGAAGAAAUCAGAGUGCUCUUUAACCCUUCAAU_UACAAUUUAUC_C_AUUCUGUUAUGCCAAAUACGUUCG ........(((..((((.((((((((((((....))))).))))))).)))))))...................................................... (-14.13 = -14.47 + 0.33)


| Location | 11,462,684 – 11,462,785 |
|---|---|
| Length | 101 |
| Sequences | 3 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 88.07 |
| Mean single sequence MFE | -25.83 |
| Consensus MFE | -16.82 |
| Energy contribution | -16.27 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.10 |
| Mean z-score | -3.08 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.714397 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11462684 101 - 27905053 AAAUUAUGGGACUUAUCGGGAAGCCAGAGUAUUUAUUAUUUAAAACUCAGUUGAUUCGGAUUUCUCAGAAAUCCUUCUGAAGAAAUCUGAGUGCUUUU--------CAA .................(..((((..((((.((((.....)))))))).....(((((((((((((((((....))))).))))))))))))))))..--------).. ( -31.70) >DroSec_CAF1 196495 109 - 1 AAAUUAUGGAACUUAUCAGGAAACCAGAGUAUAUAUUAUUUAAAACACAGUUGAUUCGGAUUUCUCAGAAAUCCUUCUGAAGAAAUCAGAGUGCUCUUUAACCCUUCAA .................(((.....(((((((.............((....))..((.((((((((((((....))))).))))))).))))))))).....))).... ( -21.10) >DroSim_CAF1 191649 109 - 1 AAAUUAUGGGACUUAUCAGGAAUCCAGAGUAUUUAUUAUUAAAAACUCAGUUUAUUCGAAUUUCUCAGAAAUUCUUCUGAAGAAAUCAGAGUGCUCUUUAACCCUUCAA .......(((................((((.((((....)))).))))((..(((((..(((((((((((....))))).))))))..)))))..))....)))..... ( -24.70) >consensus AAAUUAUGGGACUUAUCAGGAAACCAGAGUAUUUAUUAUUUAAAACUCAGUUGAUUCGGAUUUCUCAGAAAUCCUUCUGAAGAAAUCAGAGUGCUCUUUAACCCUUCAA ......(((..((....))....)))((((((.......((((.......)))).((.((((((((((((....))))).))))))).))))))))............. (-16.82 = -16.27 + -0.55)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:26:50 2006