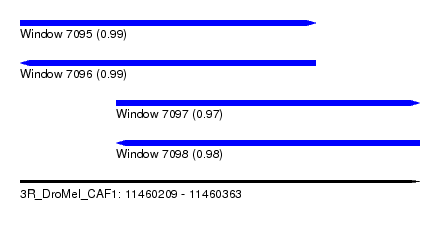
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 11,460,209 – 11,460,363 |
| Length | 154 |
| Max. P | 0.991873 |

| Location | 11,460,209 – 11,460,323 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.37 |
| Mean single sequence MFE | -39.68 |
| Consensus MFE | -33.27 |
| Energy contribution | -33.67 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.84 |
| SVM decision value | 2.24 |
| SVM RNA-class probability | 0.990924 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11460209 114 + 27905053 CAGUUUUAACCUUU-UUUUCUCUGUGUGUUGAC--GGCCAGUAAUUUGUGUUGUUGCUGUUUGUUGGUUGGCUGUCGGCUGAAGGGAAACGCCAGCAGCAAC---AACAAUGGCCACAAA ..(((...(((...-........).))...)))--(((((........(((((((((((((.((...(..((.....))..)..(....))).)))))))))---)))).)))))..... ( -35.40) >DroSec_CAF1 192736 120 + 1 CAGUUUUAACCUUUUUUUUCUCUGUGUGUUGACGUGGCCAGUAAUUUGUGUUGUUGCUGUUUGUUGGUUGGCUGUCGGCUGAAGGGAAACGCCAGCAGCAACAACAACAAUGGCCACAAA ..(((...(((............).))...)))(((((((.....((((((((((((((((.((...(..((.....))..)..(....))).)))))))))))).)))))))))))... ( -40.60) >DroSim_CAF1 187850 120 + 1 CAGUUUUAACGUUUUUUUUCUCUGUGUGUUGACGUGGCCAGUAAUUUGUGUUGUUGCUGUUUGUUGGUUGGCUGUCGGCUGAAGGGAAACGCCAGCAGCAACAACAACAAUGGCCACAAA ..(((...(((.............)))...)))(((((((.....((((((((((((((((.((...(..((.....))..)..(....))).)))))))))))).)))))))))))... ( -40.82) >DroEre_CAF1 181870 115 + 1 CAGUUGUAAC-UUU-UUUUCUCCGUGUGUUGACGUGGCCAGUAAUUUGUGUUGUUGCUGUUUGUUGGUUGGCUGUCGGCUGAAGGGAAACGCCAGCAGCAAC---AACAAUGGCCACGAA ..........-...-.................((((((((........(((((((((((((.((...(..((.....))..)..(....))).)))))))))---)))).)))))))).. ( -40.80) >DroYak_CAF1 185962 116 + 1 CAGUUGUAACCUUU-UUUUCUCUGUGUGUUGACGUGGCCAGUAAUUUGUGUUGUUGCUGUUUGUUGGUUGGCUGUCGGCUGAAGGGAAACGCCAGCAGCAAC---AACAAUGGCCACGAA ..............-.................((((((((........(((((((((((((.((...(..((.....))..)..(....))).)))))))))---)))).)))))))).. ( -40.80) >consensus CAGUUUUAACCUUU_UUUUCUCUGUGUGUUGACGUGGCCAGUAAUUUGUGUUGUUGCUGUUUGUUGGUUGGCUGUCGGCUGAAGGGAAACGCCAGCAGCAAC___AACAAUGGCCACAAA .................................(((((((.........((((((((((....(..((((.....))))..)..(....)..))))))))))........)))))))... (-33.27 = -33.67 + 0.40)



| Location | 11,460,209 – 11,460,323 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.37 |
| Mean single sequence MFE | -31.22 |
| Consensus MFE | -25.88 |
| Energy contribution | -26.04 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.53 |
| Structure conservation index | 0.83 |
| SVM decision value | 2.29 |
| SVM RNA-class probability | 0.991873 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11460209 114 - 27905053 UUUGUGGCCAUUGUU---GUUGCUGCUGGCGUUUCCCUUCAGCCGACAGCCAACCAACAAACAGCAACAACACAAAUUACUGGCC--GUCAACACACAGAGAAAA-AAAGGUUAAAACUG .(((((((((.((((---(((((((.(((((((...........))).)))).........)))))))))))........)))))--).))).............-.............. ( -26.90) >DroSec_CAF1 192736 120 - 1 UUUGUGGCCAUUGUUGUUGUUGCUGCUGGCGUUUCCCUUCAGCCGACAGCCAACCAACAAACAGCAACAACACAAAUUACUGGCCACGUCAACACACAGAGAAAAAAAAGGUUAAAACUG ...((((((((((((((((((((((.((((...........)))).)))).........)))))))))))..........)))))))................................. ( -33.60) >DroSim_CAF1 187850 120 - 1 UUUGUGGCCAUUGUUGUUGUUGCUGCUGGCGUUUCCCUUCAGCCGACAGCCAACCAACAAACAGCAACAACACAAAUUACUGGCCACGUCAACACACAGAGAAAAAAAACGUUAAAACUG ...((((((((((((((((((((((.((((...........)))).)))).........)))))))))))..........)))))))................................. ( -33.60) >DroEre_CAF1 181870 115 - 1 UUCGUGGCCAUUGUU---GUUGCUGCUGGCGUUUCCCUUCAGCCGACAGCCAACCAACAAACAGCAACAACACAAAUUACUGGCCACGUCAACACACGGAGAAAA-AAA-GUUACAACUG ..((((((((.((((---(((((((.(((((((...........))).)))).........)))))))))))........)))))))).................-...-.......... ( -31.00) >DroYak_CAF1 185962 116 - 1 UUCGUGGCCAUUGUU---GUUGCUGCUGGCGUUUCCCUUCAGCCGACAGCCAACCAACAAACAGCAACAACACAAAUUACUGGCCACGUCAACACACAGAGAAAA-AAAGGUUACAACUG ..((((((((.((((---(((((((.(((((((...........))).)))).........)))))))))))........)))))))).................-.............. ( -31.00) >consensus UUUGUGGCCAUUGUU___GUUGCUGCUGGCGUUUCCCUUCAGCCGACAGCCAACCAACAAACAGCAACAACACAAAUUACUGGCCACGUCAACACACAGAGAAAA_AAAGGUUAAAACUG ..(((((((((((((...(((((((.((((...........)))).))).)))).)))))...(......).........))))))))................................ (-25.88 = -26.04 + 0.16)


| Location | 11,460,246 – 11,460,363 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.31 |
| Mean single sequence MFE | -35.00 |
| Consensus MFE | -31.76 |
| Energy contribution | -31.52 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.02 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.67 |
| SVM RNA-class probability | 0.971158 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11460246 117 + 27905053 GUAAUUUGUGUUGUUGCUGUUUGUUGGUUGGCUGUCGGCUGAAGGGAAACGCCAGCAGCAAC---AACAAUGGCCACAAACACACAUGCAAUUUUGUGCAUUAAAAUUUCACACUGCGUU (((...((((((((.((((((.((((....(((((.(((.....(....)))).)))))..)---))))))))).)).)))))).(((((......))))).............)))... ( -35.30) >DroSec_CAF1 192776 120 + 1 GUAAUUUGUGUUGUUGCUGUUUGUUGGUUGGCUGUCGGCUGAAGGGAAACGCCAGCAGCAACAACAACAAUGGCCACAAACACACAUGCAAUUUUGUGCAUUAAAAUUUCACACUGCGUU (((...((((((((.((((((.((((....(((((.(((.....(....)))).))))).....)))))))))).)).)))))).(((((......))))).............)))... ( -35.40) >DroSim_CAF1 187890 120 + 1 GUAAUUUGUGUUGUUGCUGUUUGUUGGUUGGCUGUCGGCUGAAGGGAAACGCCAGCAGCAACAACAACAAUGGCCACAAACACACAUGCAAUUUUGUGCAUUAAAAUUUCACACUGCGUU (((...((((((((.((((((.((((....(((((.(((.....(....)))).))))).....)))))))))).)).)))))).(((((......))))).............)))... ( -35.40) >DroEre_CAF1 181908 117 + 1 GUAAUUUGUGUUGUUGCUGUUUGUUGGUUGGCUGUCGGCUGAAGGGAAACGCCAGCAGCAAC---AACAAUGGCCACGAACACACAUGCAAUUUUGUGCAUUAAAAUUUCACACUGCGUU (((...((((((((.((((((.((((....(((((.(((.....(....)))).)))))..)---))))))))).)).)))))).(((((......))))).............)))... ( -35.30) >DroYak_CAF1 186001 117 + 1 GUAAUUUGUGUUGUUGCUGUUUGUUGGUUGGCUGUCGGCUGAAGGGAAACGCCAGCAGCAAC---AACAAUGGCCACGAACACACAUGCAAUUUUGGACAUUAAAAUUUCACACUGCGUU ......((((((((.((((((.((((....(((((.(((.....(....)))).)))))..)---))))))))).)).)))))).(((((....((((.........))))...))))). ( -33.60) >consensus GUAAUUUGUGUUGUUGCUGUUUGUUGGUUGGCUGUCGGCUGAAGGGAAACGCCAGCAGCAAC___AACAAUGGCCACAAACACACAUGCAAUUUUGUGCAUUAAAAUUUCACACUGCGUU (((...((((((((.((((((.(((.((((.((((.(((.....(....)))).))))))))...))))))))).))).)))))..((.(((((((.....))))))).))...)))... (-31.76 = -31.52 + -0.24)


| Location | 11,460,246 – 11,460,363 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.31 |
| Mean single sequence MFE | -31.25 |
| Consensus MFE | -28.42 |
| Energy contribution | -28.46 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.93 |
| SVM RNA-class probability | 0.982937 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11460246 117 - 27905053 AACGCAGUGUGAAAUUUUAAUGCACAAAAUUGCAUGUGUGUUUGUGGCCAUUGUU---GUUGCUGCUGGCGUUUCCCUUCAGCCGACAGCCAACCAACAAACAGCAACAACACAAAUUAC ...((((((((...........))))...))))...((((((.((.((..(((((---(((((((.((((...........)))).))).))).))))))...)).))))))))...... ( -30.80) >DroSec_CAF1 192776 120 - 1 AACGCAGUGUGAAAUUUUAAUGCACAAAAUUGCAUGUGUGUUUGUGGCCAUUGUUGUUGUUGCUGCUGGCGUUUCCCUUCAGCCGACAGCCAACCAACAAACAGCAACAACACAAAUUAC .(((((.(((..((((((.......))))))))))))))(((((((....(((((((((((((((.((((...........)))).)))).........))))))))))))))))))... ( -32.60) >DroSim_CAF1 187890 120 - 1 AACGCAGUGUGAAAUUUUAAUGCACAAAAUUGCAUGUGUGUUUGUGGCCAUUGUUGUUGUUGCUGCUGGCGUUUCCCUUCAGCCGACAGCCAACCAACAAACAGCAACAACACAAAUUAC .(((((.(((..((((((.......))))))))))))))(((((((....(((((((((((((((.((((...........)))).)))).........))))))))))))))))))... ( -32.60) >DroEre_CAF1 181908 117 - 1 AACGCAGUGUGAAAUUUUAAUGCACAAAAUUGCAUGUGUGUUCGUGGCCAUUGUU---GUUGCUGCUGGCGUUUCCCUUCAGCCGACAGCCAACCAACAAACAGCAACAACACAAAUUAC ...((((((((...........))))...))))...((((((.((.((..(((((---(((((((.((((...........)))).))).))).))))))...)).))))))))...... ( -30.80) >DroYak_CAF1 186001 117 - 1 AACGCAGUGUGAAAUUUUAAUGUCCAAAAUUGCAUGUGUGUUCGUGGCCAUUGUU---GUUGCUGCUGGCGUUUCCCUUCAGCCGACAGCCAACCAACAAACAGCAACAACACAAAUUAC ...(((((....................)))))...((((((.((.((..(((((---(((((((.((((...........)))).))).))).))))))...)).))))))))...... ( -29.45) >consensus AACGCAGUGUGAAAUUUUAAUGCACAAAAUUGCAUGUGUGUUUGUGGCCAUUGUU___GUUGCUGCUGGCGUUUCCCUUCAGCCGACAGCCAACCAACAAACAGCAACAACACAAAUUAC ...((((((((...........))))...))))...((((((.((.((..(((((...(((((((.((((...........)))).))).)))).)))))...)).))))))))...... (-28.42 = -28.46 + 0.04)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:26:45 2006