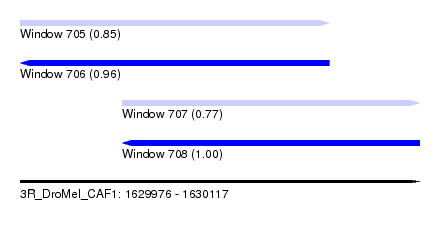
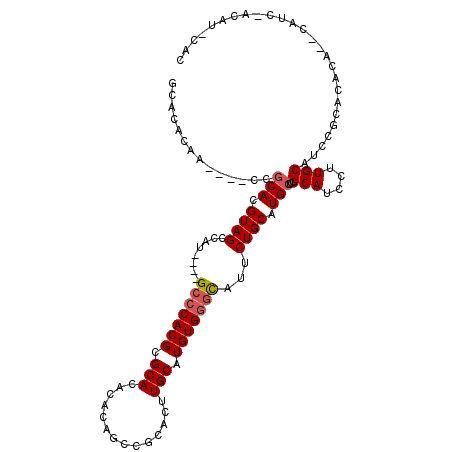
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 1,629,976 – 1,630,117 |
| Length | 141 |
| Max. P | 0.998276 |

| Location | 1,629,976 – 1,630,085 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 81.47 |
| Mean single sequence MFE | -32.18 |
| Consensus MFE | -24.48 |
| Energy contribution | -24.92 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.78 |
| SVM RNA-class probability | 0.848353 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 1629976 109 + 27905053 UUCUAGCUGAUUUGUCAUGUGGAUGUGGAUGUAGUUGUGAAUGUCGAUG--UGUGUGUGGAUGCAAGGAUGCGAGCAUGCAGAAUGCCCACAUGCAAGUGCGGCUGUGUGU ...(((((((((((.(((((((((((.(((((........))))).)))--)((((((...((((....)))).)))))).......)))))))))))).))))))..... ( -35.50) >DroSec_CAF1 9295 111 + 1 UUCUAGCUGAUUUGUCAUGUGGAUGUGGAUGUAGAUGUGGAUGUAGAUGAGUGUGUGCGGAUGCAGGGAUGCGAGCAUGCAGAAUGCCCACAUGCAAGUGCGGCUGUGUGU ...(((((((((((.(((((((.......((((..((((..((((.((....)).))))..))))....)))).((((.....)))))))))))))))).))))))..... ( -35.60) >DroSim_CAF1 9299 111 + 1 UUCUAGCUGAUUUGUCAUGUGGAUGUGGAUGUAGAUGUGGAUGUGGAUGAGUGUGUGCGGAUGCAAGGAUGCGAGCAUGCAGAAUGCCCACAUGCAAGUGCGGCUGUGUGU ...(((((((((((.(((((((.......((((....((.((.((.((......)).)).)).))....)))).((((.....)))))))))))))))).))))))..... ( -34.20) >DroEre_CAF1 9445 85 + 1 UUCUAGCUGAUUUGUCAUGUGGAUGUGGAUGUGGC--------------------------UGCAAGGAUGCCAGCAUGCAGAAUACCCACAUGCAAGUACGCCUGUGUGU .....((..(((((.(((((((.(((...((((((--------------------------((((....)).)))).))))..))).))))))))))))..))........ ( -26.30) >DroYak_CAF1 8676 90 + 1 UUCUAGCUGAUUUGUCAUGUGGAUGUGGAUGUAGC---------------------AAGGAUGCAAGGAUGCCAGCAUGCAGAAUGUCCACAAGCAAGUGCGGCUGUGUGU ...((((((((((((..(((((((((...((((((---------------------..(.((......)).)..)).))))..))))))))).)))))).))))))..... ( -29.30) >consensus UUCUAGCUGAUUUGUCAUGUGGAUGUGGAUGUAGAUGUG_AUGU_GAUG__UGUGUGCGGAUGCAAGGAUGCGAGCAUGCAGAAUGCCCACAUGCAAGUGCGGCUGUGUGU ...(((((((((((.(((((((........................................(((....)))..((((.....)))))))))))))))).))))))..... (-24.48 = -24.92 + 0.44)

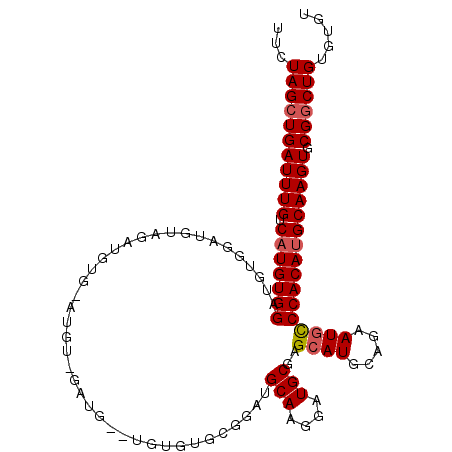
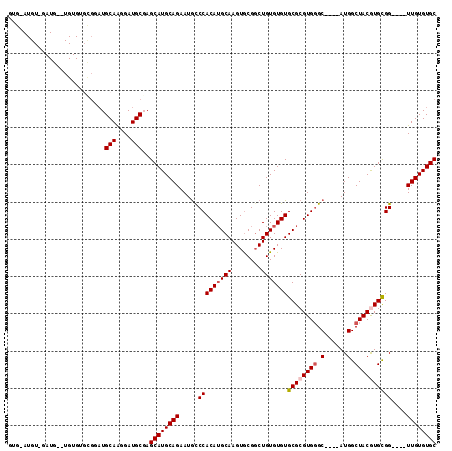
| Location | 1,629,976 – 1,630,085 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 81.47 |
| Mean single sequence MFE | -20.32 |
| Consensus MFE | -16.70 |
| Energy contribution | -17.14 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.57 |
| SVM RNA-class probability | 0.964750 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 1629976 109 - 27905053 ACACACAGCCGCACUUGCAUGUGGGCAUUCUGCAUGCUCGCAUCCUUGCAUCCACACACA--CAUCGACAUUCACAACUACAUCCACAUCCACAUGACAAAUCAGCUAGAA ......(((.(...((((((((((((((.....))))..(((....)))...........--...........................))))))).)))..).))).... ( -20.20) >DroSec_CAF1 9295 111 - 1 ACACACAGCCGCACUUGCAUGUGGGCAUUCUGCAUGCUCGCAUCCCUGCAUCCGCACACACUCAUCUACAUCCACAUCUACAUCCACAUCCACAUGACAAAUCAGCUAGAA ......(((.(...((((((((((((((.....))))..(((....)))........................................))))))).)))..).))).... ( -19.40) >DroSim_CAF1 9299 111 - 1 ACACACAGCCGCACUUGCAUGUGGGCAUUCUGCAUGCUCGCAUCCUUGCAUCCGCACACACUCAUCCACAUCCACAUCUACAUCCACAUCCACAUGACAAAUCAGCUAGAA ......(((.(...((((((((((((((.....))))..(((....)))........................................))))))).)))..).))).... ( -20.20) >DroEre_CAF1 9445 85 - 1 ACACACAGGCGUACUUGCAUGUGGGUAUUCUGCAUGCUGGCAUCCUUGCA--------------------------GCCACAUCCACAUCCACAUGACAAAUCAGCUAGAA .......(((....((((((((((((....((.(((.((((.........--------------------------))))))).)).))))))))).)))....))).... ( -24.30) >DroYak_CAF1 8676 90 - 1 ACACACAGCCGCACUUGCUUGUGGACAUUCUGCAUGCUGGCAUCCUUGCAUCCUU---------------------GCUACAUCCACAUCCACAUGACAAAUCAGCUAGAA ......(((.((....)).((((((......(((.....(((....))).....)---------------------))....))))))................))).... ( -17.50) >consensus ACACACAGCCGCACUUGCAUGUGGGCAUUCUGCAUGCUCGCAUCCUUGCAUCCGCACACA__CAUC_ACAU_CACAGCUACAUCCACAUCCACAUGACAAAUCAGCUAGAA ......(((.(...((((((((((((((.....))))..(((....)))........................................))))))).)))..).))).... (-16.70 = -17.14 + 0.44)

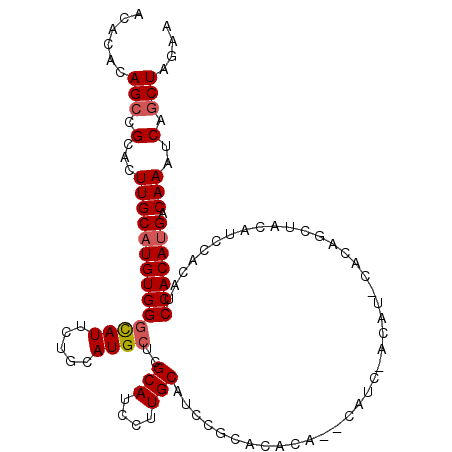
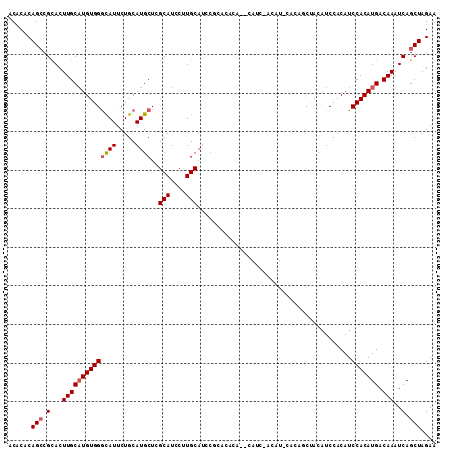
| Location | 1,630,012 – 1,630,117 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 78.76 |
| Mean single sequence MFE | -36.94 |
| Consensus MFE | -26.50 |
| Energy contribution | -27.14 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.773765 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 1630012 105 + 27905053 GUGAAUGUCGAUG--UGUGUGUGGAUGCAAGGAUGCGAGCAUGCAGAAUGCCCACAUGCAAGUGCGGCUGUGUGUGCGCGUGGGC----AUGGCUACGUGCGG----UUGUGUGC ......((((...--(((((((((.((((....)))).((((.....)))))))))))))....)))).....(..((((...((----(((....)))))..----.))))..) ( -37.50) >DroSec_CAF1 9331 107 + 1 GUGGAUGUAGAUGAGUGUGUGCGGAUGCAGGGAUGCGAGCAUGCAGAAUGCCCACAUGCAAGUGCGGCUGUGUGUGCGCGUGGGC----AUGGCUACGUGCGG----UUGUGUGC .((.((((((.....(((((((...((((....)))).)))))))..((((((((.((((.(..(....)..).)))).))))))----))..)))))).)).----........ ( -40.90) >DroSim_CAF1 9335 107 + 1 GUGGAUGUGGAUGAGUGUGUGCGGAUGCAAGGAUGCGAGCAUGCAGAAUGCCCACAUGCAAGUGCGGCUGUGUGUGCGCGUGGGC----AUGGCUACGUGCGG----UUGUGUGC .((.((((((.....(((((((...((((....)))).)))))))..((((((((.((((.(..(....)..).)))).))))))----))..)))))).)).----........ ( -39.80) >DroEre_CAF1 9480 82 + 1 -------------------------UGCAAGGAUGCCAGCAUGCAGAAUACCCACAUGCAAGUACGCCUGUGUGUGCGCGUGGGC----AUGGCUACAUGCGG----UUGUGUGC -------------------------.(((.....(((.(((((.((.((.(((((.((((..(((....)))..)))).))))).----))..)).)))))))----)....))) ( -30.40) >DroYak_CAF1 8711 95 + 1 --------------------AAGGAUGCAAGGAUGCCAGCAUGCAGAAUGUCCACAAGCAAGUGCGGCUGUGUGUGCGCGUGUGCAUGCAUGGCUACAUGUGGUUGGUUGUGUGC --------------------......(((....)))..((((((((.....(((((.((....))(((((((((((((....)))))))))))))...)))))....)))))))) ( -36.10) >consensus GUG_AUGU_GAUG__UGUGUGCGGAUGCAAGGAUGCGAGCAUGCAGAAUGCCCACAUGCAAGUGCGGCUGUGUGUGCGCGUGGGC____AUGGCUACGUGCGG____UUGUGUGC ..........................(((....)))..((((((((.....(((((((((........)))))))((((((((.(......).))))))))))....)))))))) (-26.50 = -27.14 + 0.64)

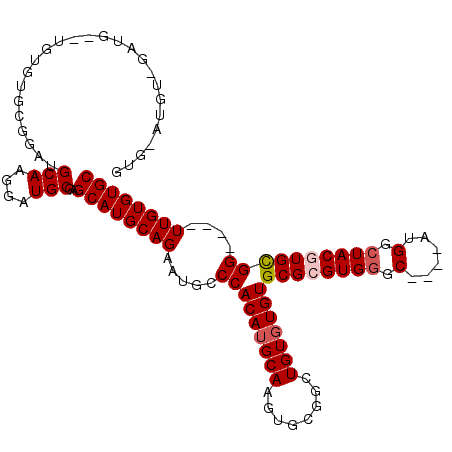
| Location | 1,630,012 – 1,630,117 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 78.76 |
| Mean single sequence MFE | -29.42 |
| Consensus MFE | -18.86 |
| Energy contribution | -19.30 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.05 |
| Mean z-score | -3.06 |
| Structure conservation index | 0.64 |
| SVM decision value | 3.05 |
| SVM RNA-class probability | 0.998276 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 1630012 105 - 27905053 GCACACAA----CCGCACGUAGCCAU----GCCCACGCGCACACACAGCCGCACUUGCAUGUGGGCAUUCUGCAUGCUCGCAUCCUUGCAUCCACACACA--CAUCGACAUUCAC ........----..(((.((((..((----(((((((.((.......)))((....))..)))))))).)))).)))..(((....)))...........--............. ( -28.20) >DroSec_CAF1 9331 107 - 1 GCACACAA----CCGCACGUAGCCAU----GCCCACGCGCACACACAGCCGCACUUGCAUGUGGGCAUUCUGCAUGCUCGCAUCCCUGCAUCCGCACACACUCAUCUACAUCCAC ((......----..(((.((((..((----(((((((.((.......)))((....))..)))))))).)))).)))..(((....)))....)).................... ( -29.90) >DroSim_CAF1 9335 107 - 1 GCACACAA----CCGCACGUAGCCAU----GCCCACGCGCACACACAGCCGCACUUGCAUGUGGGCAUUCUGCAUGCUCGCAUCCUUGCAUCCGCACACACUCAUCCACAUCCAC ((......----..(((.((((..((----(((((((.((.......)))((....))..)))))))).)))).)))..(((....)))....)).................... ( -30.70) >DroEre_CAF1 9480 82 - 1 GCACACAA----CCGCAUGUAGCCAU----GCCCACGCGCACACACAGGCGUACUUGCAUGUGGGUAUUCUGCAUGCUGGCAUCCUUGCA------------------------- (((.....----((((((((((..((----(((((((.(((...((....))...))).))))))))).)))))))).))......))).------------------------- ( -32.60) >DroYak_CAF1 8711 95 - 1 GCACACAACCAACCACAUGUAGCCAUGCAUGCACACGCGCACACACAGCCGCACUUGCUUGUGGACAUUCUGCAUGCUGGCAUCCUUGCAUCCUU-------------------- (((.(((..........))).((((.(((((((.....(..((((.(((.......)))))))..)....))))))))))).....)))......-------------------- ( -25.70) >consensus GCACACAA____CCGCACGUAGCCAU____GCCCACGCGCACACACAGCCGCACUUGCAUGUGGGCAUUCUGCAUGCUCGCAUCCUUGCAUCCGCACACA__CAUC_ACAU_CAC ..............(((.((((........(((((((.(((..............))).)))))))...)))).)))..(((....))).......................... (-18.86 = -19.30 + 0.44)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:43:06 2006