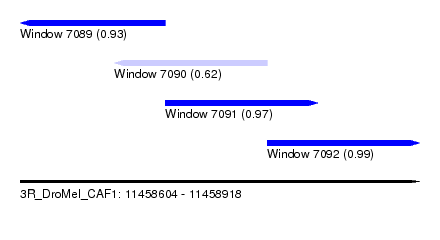
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 11,458,604 – 11,458,918 |
| Length | 314 |
| Max. P | 0.992572 |

| Location | 11,458,604 – 11,458,718 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.56 |
| Mean single sequence MFE | -32.75 |
| Consensus MFE | -29.50 |
| Energy contribution | -29.70 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.18 |
| SVM RNA-class probability | 0.926351 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11458604 114 - 27905053 ACGCAUGGCAACAAUUGCAGCCAGUUUAAUGAGCUCAAAAAUGCGCGCAACAAUGAAGAGCUGGCCCGGCUCCAAACCAACGCCAAUUCC------CCGAUGGCCAACCCUACUCGGCAU ..(((((....))..))).((((((((.....((.((....)).)).((....))..))))))))..(((...........)))......------(((((((......))).))))... ( -25.50) >DroSec_CAF1 191121 114 - 1 ACGCAUGGCAACAAUUGCAGCCGGUUUAAUGAGCUCAAAAAUGCGCGCAACAAUGAAGAGCUGGCCCGGCUCCAAACCAAGGCCAAUUCC------CCGUUGGCCAACCCGACUCGGCAU ..(((((....))..))).((((((((..((.((.((....)).)).))........((((((...)))))).)))))..(((((((...------..)))))))..........))).. ( -36.80) >DroSim_CAF1 186239 114 - 1 ACGCAUGGCAACAAUUGCAGCCGGUUUAAUGAGCUCAAAAAUGCGCGCAACAAUGAAGAGCUGGCCCGGCUCCAAACCAAGGCCAAUUCC------CCGAUGGCCAACCCCACUCGGCAU ..(((((....))..))).((((((((..((.((.((....)).)).))........((((((...)))))).)))))..(((((.....------....)))))..........))).. ( -33.90) >DroEre_CAF1 180347 120 - 1 ACGCAUGGCAACAAUUGCAGCCGGUUUAAUGAGCUCAAAAAUGCGCGCAACAAUGAAGAGCUGGCCGAGCUCCAAGCCAAGGCCAAUUCCCAGUCCCAAAUGGCCAACCCCACGCGGCAU ..(((((....))..))).((((((((...((((((......((...((....))....)).....)))))).)))))..(((((...............)))))..........))).. ( -34.06) >DroYak_CAF1 184364 114 - 1 ACGCAUGGCAACAAUUGCAGCCAGUUUAAUGAGCUCAAAAAUGCGCGCAACAAUGAAGAGCUGGCCCGGCUCCAAGCCAAGGCCAAUUCC------CCAAUGGCCAACCCCACGCGGCAU ..(((((....))..))).(((..........((........))(((......((..(((.(((((.(((.....)))..))))).))).------.)).(((......))))))))).. ( -33.50) >consensus ACGCAUGGCAACAAUUGCAGCCGGUUUAAUGAGCUCAAAAAUGCGCGCAACAAUGAAGAGCUGGCCCGGCUCCAAACCAAGGCCAAUUCC______CCGAUGGCCAACCCCACUCGGCAU ..(((((....))..))).((((((((..((.((.((....)).)).))........(((((.....))))).)))))..(((((...............)))))..........))).. (-29.50 = -29.70 + 0.20)

| Location | 11,458,678 – 11,458,798 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.29 |
| Mean single sequence MFE | -32.34 |
| Consensus MFE | -27.84 |
| Energy contribution | -27.80 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.31 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.616888 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11458678 120 - 27905053 GACUGUCGACUGCUGACUGCUGACUGCAAAUUGAUUGCCGGCCAACAAAAGGGCAGAAACUGCAGCAGAGUACUCACGCAACGCAUGGCAACAAUUGCAGCCAGUUUAAUGAGCUCAAAA ...........(((.(..((((.((((((.(((.((((((((..........((((...)))).((.((....))..))...)).))))))))))))))).))))....).)))...... ( -33.00) >DroSec_CAF1 191195 113 - 1 GACUGUCGA-------CUGCUGACUGCAAAUUGAUUGCCGGCAAACAAAAGGGCAGAAUCUGCAGCAGAGUACUCACGCAACGCAUGGCAACAAUUGCAGCCGGUUUAAUGAGCUCAAAA .....(((.-------..((((.((((((.(((.((((((((..........((((...)))).((.((....))..))...)).))))))))))))))).))))....)))........ ( -30.70) >DroSim_CAF1 186313 120 - 1 GACUGUCGACUCCUGACUGCUGACUGCAAAUUGAUUGCCGGCCAACAAAAGGGCAGAAUCUGCAGCAGAGUACUCACGCAACGCAUGGCAACAAUUGCAGCCGGUUUAAUGAGCUCAAAA .......(((((......((((.((((((.(((.((((((((..........((((...)))).((.((....))..))...)).))))))))))))))).)))).....))).)).... ( -33.10) >DroEre_CAF1 180427 106 - 1 --------------GUCUGCUGACUGCAAAUUGAUUGCCGGCCAACAAAAGGGCAGAAACUGCAGCGGAGUACUCACGCAACGCAUGGCAACAAUUGCAGCCGGUUUAAUGAGCUCAAAA --------------....((((.((((((.(((.((((((((..........((((...)))).(((((....)).)))...)).))))))))))))))).))))............... ( -32.80) >DroYak_CAF1 184438 120 - 1 AACUGCAGACUGUCGACUGCUGACUGCAAAUUGAUUGCCGGGCAACAAAAGGGCAGAAACUGCAGCAGAGUACUCACGCAACGCAUGGCAACAAUUGCAGCCAGUUUAAUGAGCUCAAAA .......((...(((...((((.((((((.(((.((((((.((.........((((...)))).((.((....))..))...)).))))))))))))))).))))....)))..)).... ( -32.10) >consensus GACUGUCGACU___GACUGCUGACUGCAAAUUGAUUGCCGGCCAACAAAAGGGCAGAAACUGCAGCAGAGUACUCACGCAACGCAUGGCAACAAUUGCAGCCGGUUUAAUGAGCUCAAAA ..................((((.((((((.(((.((((((((..........((((...)))).((.((....))..))...)).))))))))))))))).))))............... (-27.84 = -27.80 + -0.04)


| Location | 11,458,718 – 11,458,838 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.79 |
| Mean single sequence MFE | -37.16 |
| Consensus MFE | -28.94 |
| Energy contribution | -29.46 |
| Covariance contribution | 0.52 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.46 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.67 |
| SVM RNA-class probability | 0.970899 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11458718 120 + 27905053 UGCGUGAGUACUCUGCUGCAGUUUCUGCCCUUUUGUUGGCCGGCAAUCAAUUUGCAGUCAGCAGUCAGCAGUCGACAGUCUGCAGUUUGCAAAAGUUGCAGAAAUGCAAAAACGAAAUUA (((((..((...(((((((((...))))....((((((((..((((.....)))).))))))))..)))))...))..((((((..((....))..)))))).)))))............ ( -37.90) >DroSec_CAF1 191235 113 + 1 UGCGUGAGUACUCUGCUGCAGAUUCUGCCCUUUUGUUUGCCGGCAAUCAAUUUGCAGUCAGCAG-------UCGACAGUCUGCAGUUUGCAAAAGUUGCAGAAAUGCAAAAACGAAAUUA (((....)))......((((..((((((.(((((((..((..((((.....)))).....((((-------.(....).)))).))..)))))))..)))))).))))............ ( -34.90) >DroSim_CAF1 186353 120 + 1 UGCGUGAGUACUCUGCUGCAGAUUCUGCCCUUUUGUUGGCCGGCAAUCAAUUUGCAGUCAGCAGUCAGGAGUCGACAGUCUGCAGUUUGCAAAAGUUGCAGAAAUGCAAAAACGAAAUUA ..(((..(((....(((((((((((.(((((.((((((((..((((.....)))).))))))))..))).)).))..))))))))).))).....(((((....)))))..)))...... ( -41.70) >DroEre_CAF1 180467 106 + 1 UGCGUGAGUACUCCGCUGCAGUUUCUGCCCUUUUGUUGGCCGGCAAUCAAUUUGCAGUCAGCAGAC--------------UGCAGUUUGCAAAAGUUGCAGAAAUGCAAAAACGAAAUUA .(((.((....)))))(((((...((((...(((((((((..((((.....)))).))))))))).--------------.)))).)))))....(((((....)))))........... ( -35.70) >DroYak_CAF1 184478 120 + 1 UGCGUGAGUACUCUGCUGCAGUUUCUGCCCUUUUGUUGCCCGGCAAUCAAUUUGCAGUCAGCAGUCGACAGUCUGCAGUUUGCAGUUUGCAAAAGUUGCAGAAAUGCAAAAACGAAAUUA (((....)))......((((.(((((((.((((((((((...)))).(((.((((((...((((.(....).))))...)))))).)))))))))..)))))))))))............ ( -35.60) >consensus UGCGUGAGUACUCUGCUGCAGUUUCUGCCCUUUUGUUGGCCGGCAAUCAAUUUGCAGUCAGCAGUC___AGUCGACAGUCUGCAGUUUGCAAAAGUUGCAGAAAUGCAAAAACGAAAUUA .(((.((....)))))((((.(((((((.(((((((..((..((((.....)))).....((((...............)))).))..)))))))..)))))))))))............ (-28.94 = -29.46 + 0.52)

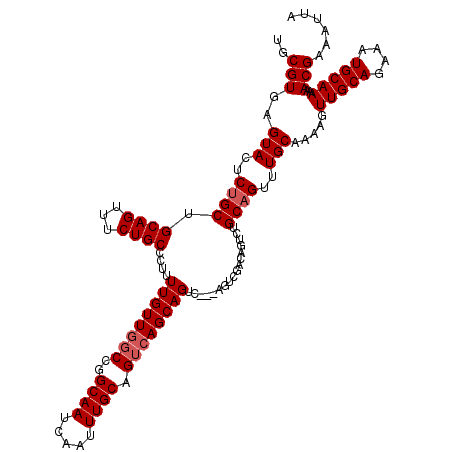
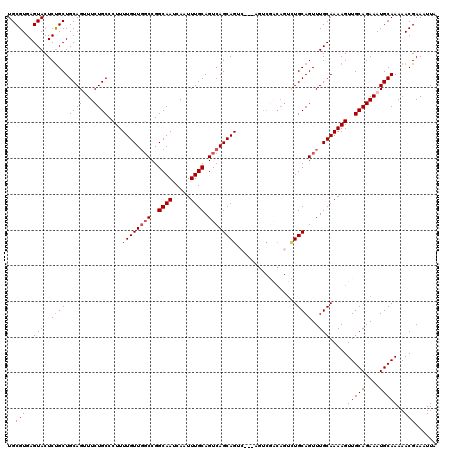
| Location | 11,458,798 – 11,458,918 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.17 |
| Mean single sequence MFE | -31.50 |
| Consensus MFE | -27.60 |
| Energy contribution | -29.80 |
| Covariance contribution | 2.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.43 |
| Structure conservation index | 0.88 |
| SVM decision value | 2.34 |
| SVM RNA-class probability | 0.992572 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11458798 120 + 27905053 UGCAGUUUGCAAAAGUUGCAGAAAUGCAAAAACGAAAUUAUAUUUGCGGGGCCCAGGCACAUCAUUAAAAUGCAUCGACCUGCAGAACAAACGAGUCGAACACCUCUCGGCAAAAAGCUU .((..(((((.....(((((....))))).............((((((((......(((...........))).....)))))))).....((((..(....)..)))))))))..)).. ( -31.50) >DroSec_CAF1 191308 120 + 1 UGCAGUUUGCAAAAGUUGCAGAAAUGCAAAAACGAAAUUAUAUUUGCGGGGCCCAGGCACAUCAUUAAAAUGCAUCGACCUGCAGAACAAACGAGUCGAACACCUCUCGGCAAAAAGCUU .((..(((((.....(((((....))))).............((((((((......(((...........))).....)))))))).....((((..(....)..)))))))))..)).. ( -31.50) >DroSim_CAF1 186433 120 + 1 UGCAGUUUGCAAAAGUUGCAGAAAUGCAAAAACGAAAUUAUAUUUGCGGGGCCCAGGCACAUCAUUAAAAUGCAUCGACCUGCAGAACAAACGAGUCGAACACCUCUCGGCAAAAAGCUU .((..(((((.....(((((....))))).............((((((((......(((...........))).....)))))))).....((((..(....)..)))))))))..)).. ( -31.50) >DroEre_CAF1 180533 120 + 1 UGCAGUUUGCAAAAGUUGCAGAAAUGCAAAAACGAAAUUAUAUUUGCGGGGCCCAGGCACAUCAUUAAAAUGCAUCGACCUGCAGAGCAAACAAGUCGAACACCUCUCGGCAUAAAGCCU .((.((((((.....(((((....))))).............((((((((......(((...........))).....))))))))))))))..(((((.......))))).....)).. ( -34.90) >DroYak_CAF1 184558 120 + 1 UGCAGUUUGCAAAAGUUGCAGAAAUGCAAAAACGAAAUUAUAUUUGCGGGGCCCAGGCACAUCAUUAAAAUGCAUCGACCUGCAGAACAAACGAGUCAAACACCUCUCGGCAUAAAGCUU .((..(.(((.....(((((....)))))...(((.......((((((((......(((...........))).....))))))))......(((........))))))))).)..)).. ( -28.10) >consensus UGCAGUUUGCAAAAGUUGCAGAAAUGCAAAAACGAAAUUAUAUUUGCGGGGCCCAGGCACAUCAUUAAAAUGCAUCGACCUGCAGAACAAACGAGUCGAACACCUCUCGGCAAAAAGCUU .((.(((((......(((((....))))).............((((((((......(((...........))).....)))))))).)))))..(((((.......))))).....)).. (-27.60 = -29.80 + 2.20)


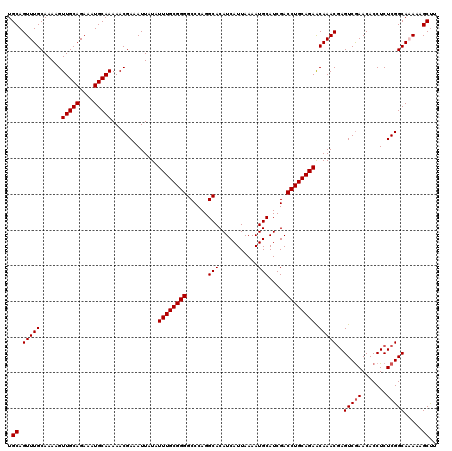
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:26:39 2006