
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 11,454,798 – 11,454,997 |
| Length | 199 |
| Max. P | 0.766379 |

| Location | 11,454,798 – 11,454,917 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.00 |
| Mean single sequence MFE | -28.82 |
| Consensus MFE | -22.94 |
| Energy contribution | -22.30 |
| Covariance contribution | -0.64 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.766379 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11454798 119 + 27905053 GCUGUCUUUGAUUUUUUUCUAAAAGCAUUUGCACUGAGUUCCUUUUU-GGGGGACGAAUUUCGUUGGAAAGUGAAUUUGGUGAGGUUUCCUGUUUUAAAAUUAGUGAUUGUGACUAAUUA ((.(((.((((((((...(...((((.....(((..((((((((((.-.(.(((.....))).)..))))).)))))..)))..))))...)....)))))))).))).))......... ( -28.60) >DroSec_CAF1 187309 118 + 1 GCUGUCUUUGAUUU-UUUCUAAAAGCAUUUACACUGAGUUCCUUUUU-GGGGGACGAAUUUCGUUGGAAAGUGAAUUUGGUGAGGUUUCCUGUUUGAAAAUUAGUGAUUGUGACUAAUUA ((.(((.(((((((-((.....((((.....(((..((((((((((.-.(.(((.....))).)..))))).)))))..)))(((...)))))))))))))))).))).))......... ( -29.40) >DroSim_CAF1 182361 118 + 1 GCUGUCUUUGAUUU-UUUCUAAAAGCAUUUACACUGAGUUCCUUUUU-GGGGGACGAAUUUCGUUGGAAAGUGAAUUUGGUGAGGUUUCCUGUUUGAAAAUUAGUGAUUGUGACUAAUUA ((.(((.(((((((-((.....((((.....(((..((((((((((.-.(.(((.....))).)..))))).)))))..)))(((...)))))))))))))))).))).))......... ( -29.40) >DroEre_CAF1 176632 112 + 1 ---GUCUUUGGUU---UUCUAAAAGCAUU-UCUCCGAGUCCCUUUUU-GGGAGGCGAAUUUCGUUGGAAAGUGAAUUUGGAGAGGCUUCCUGUUUAAAAAUUAGUGAUUGUGACUAAUUA ---......(.((---(((((((..((((-(.(((...((((.....-))))((((.....))))))))))))..))))))))).)............(((((((.......))))))). ( -28.30) >DroYak_CAF1 180381 118 + 1 GCUGUCUUUGGUU--UUUCUAAAAGCAUUUUCCCCGAGUUCCUUUUUCGGGAGACGAAUUUCGUUGGAAAGUGAAUUCGGUGAGGUUUCCUGUUUAAAAAUUAGUGAUUGUGACUAAUUA ..((..(((((..--...)))))..)).((((.(((((((((((((.((..(......)..))...))))).)))))))).)))).............(((((((.......))))))). ( -28.40) >consensus GCUGUCUUUGAUUU_UUUCUAAAAGCAUUUACACUGAGUUCCUUUUU_GGGGGACGAAUUUCGUUGGAAAGUGAAUUUGGUGAGGUUUCCUGUUUAAAAAUUAGUGAUUGUGACUAAUUA ......................((((.....((((((((((((((((.(.((((....)))).).)))))).))))))))))(((...)))))))...(((((((.......))))))). (-22.94 = -22.30 + -0.64)



| Location | 11,454,838 – 11,454,957 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.64 |
| Mean single sequence MFE | -18.74 |
| Consensus MFE | -17.38 |
| Energy contribution | -17.18 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.10 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.578924 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11454838 119 - 27905053 AGCAAAUCCCACUAAUUAUGUGAGUUUGGCAACGACAGCAUAAUUAGUCACAAUCACUAAUUUUAAAACAGGAAACCUCACCAAAUUCACUUUCCAACGAAAUUCGUCCCCC-AAAAAGG ........((.........((((((((((............(((((((.......)))))))........(....)....))))))))))......(((.....))).....-.....)) ( -18.60) >DroSec_CAF1 187348 119 - 1 GGCAAAUCCCACUAAUUGUGUGAGUUUGGCAACGACAGCAUAAUUAGUCACAAUCACUAAUUUUCAAACAGGAAACCUCACCAAAUUCACUUUCCAACGAAAUUCGUCCCCC-AAAAAGG (((.......(((((((((((....(((....)))..)))))))))))......................(((((...............)))))..........)))....-....... ( -18.96) >DroSim_CAF1 182400 119 - 1 GGCAAAUCCAACUAAUUGUGUGAGUUUGGCAACGACAGCAUAAUUAGUCACAAUCACUAAUUUUCAAACAGGAAACCUCACCAAAUUCACUUUCCAACGAAAUUCGUCCCCC-AAAAAGG (((.......(((((((((((....(((....)))..)))))))))))......................(((((...............)))))..........)))....-....... ( -18.96) >DroEre_CAF1 176665 118 - 1 U-CCAAUCCCACUAAUUAUGUGCGUUUGGCAACGACAGCAUAAUUAGUCACAAUCACUAAUUUUUAAACAGGAAGCCUCUCCAAAUUCACUUUCCAACGAAAUUCGCCUCCC-AAAAAGG .-......(((((((((((((..(((.(....)))).)))))))))))......................(((......)))..............................-.....)) ( -18.00) >DroYak_CAF1 180419 120 - 1 AGCAAAUCCCACUAAUUAUGUGAGUUUGGCAACGACAGCAUAAUUAGUCACAAUCACUAAUUUUUAAACAGGAAACCUCACCGAAUUCACUUUCCAACGAAAUUCGUCUCCCGAAAAAGG ........(((((((((((((....(((....)))..)))))))))))............(((((.....(((((...............))))).(((.....))).....))))).)) ( -19.16) >consensus AGCAAAUCCCACUAAUUAUGUGAGUUUGGCAACGACAGCAUAAUUAGUCACAAUCACUAAUUUUCAAACAGGAAACCUCACCAAAUUCACUUUCCAACGAAAUUCGUCCCCC_AAAAAGG ..........(((((((((((....(((....)))..)))))))))))......................(((((...............))))).(((.....)))............. (-17.38 = -17.18 + -0.20)

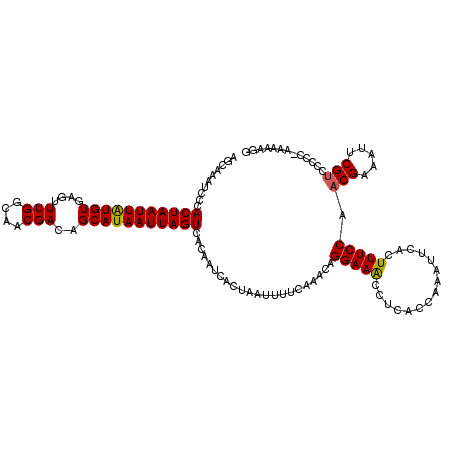
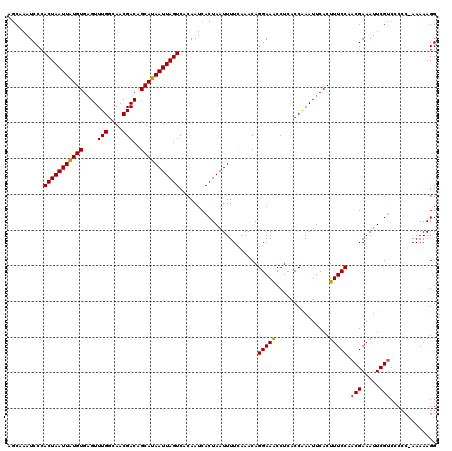
| Location | 11,454,877 – 11,454,997 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.00 |
| Mean single sequence MFE | -18.82 |
| Consensus MFE | -14.64 |
| Energy contribution | -14.40 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.559189 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11454877 120 - 27905053 CGUAUCGAGCUUACUCCACAUCAAAGCAAAUUACUCCACAAGCAAAUCCCACUAAUUAUGUGAGUUUGGCAACGACAGCAUAAUUAGUCACAAUCACUAAUUUUAAAACAGGAAACCUCA ......(((.((..(((........((..............)).......(((((((((((....(((....)))..)))))))))))......................))))).))). ( -19.54) >DroSec_CAF1 187387 120 - 1 CGUAUCGAGCUUACUCCACAUCAAAGCAAAUUACUCCACAGGCAAAUCCCACUAAUUGUGUGAGUUUGGCAACGACAGCAUAAUUAGUCACAAUCACUAAUUUUCAAACAGGAAACCUCA .(((....((((...........))))....)))(((...((......))(((((((((((....(((....)))..)))))))))))......................)))....... ( -21.00) >DroSim_CAF1 182439 120 - 1 CGUAUCGAGCUUACUCCACAUCAAAGCAAAUUACUCCACAGGCAAAUCCAACUAAUUGUGUGAGUUUGGCAACGACAGCAUAAUUAGUCACAAUCACUAAUUUUCAAACAGGAAACCUCA .(((....((((...........))))....)))(((...((.....)).(((((((((((....(((....)))..)))))))))))......................)))....... ( -20.60) >DroEre_CAF1 176704 101 - 1 ------------------CAUCAAAGCAAAGUACUCCACCU-CCAAUCCCACUAAUUAUGUGCGUUUGGCAACGACAGCAUAAUUAGUCACAAUCACUAAUUUUUAAACAGGAAGCCUCU ------------------.......((..............-........(((((((((((..(((.(....)))).)))))))))))............(((((....))))))).... ( -14.70) >DroYak_CAF1 180459 120 - 1 CAGAUCCAGCGCCCUCCACAUCAAAGCAAAUUACUCCACAAGCAAAUCCCACUAAUUAUGUGAGUUUGGCAACGACAGCAUAAUUAGUCACAAUCACUAAUUUUUAAACAGGAAACCUCA ....(((.((...............)).......................(((((((((((....(((....)))..)))))))))))......................)))....... ( -18.26) >consensus CGUAUCGAGCUUACUCCACAUCAAAGCAAAUUACUCCACAAGCAAAUCCCACUAAUUAUGUGAGUUUGGCAACGACAGCAUAAUUAGUCACAAUCACUAAUUUUCAAACAGGAAACCUCA ..................................................(((((((((((....(((....)))..)))))))))))......................(....).... (-14.64 = -14.40 + -0.24)


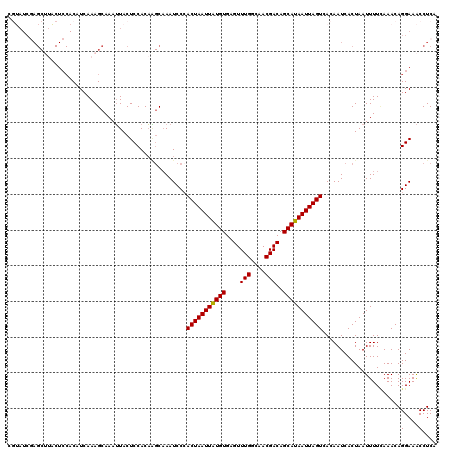
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:26:29 2006