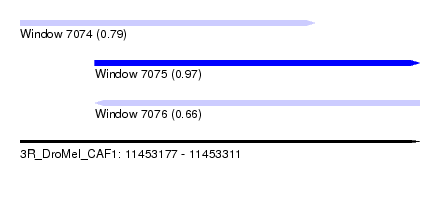
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 11,453,177 – 11,453,311 |
| Length | 134 |
| Max. P | 0.967285 |

| Location | 11,453,177 – 11,453,276 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.41 |
| Mean single sequence MFE | -35.50 |
| Consensus MFE | -21.43 |
| Energy contribution | -21.35 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.785628 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11453177 99 + 27905053 ACACACCGUGCACUAGCUCCU---------------CGAAUCCAAA----GCGAAAAUCUGCGCAUAAGCAGCUUGACUCUUGCCAGAUUGGAUGCGAUGGAGGUG--GGCAAUCCUGCC ........(((.(((.(((((---------------((.(((((..----......(((((.(((..(((.....).))..))))))))))))).))).)))).))--))))........ ( -32.00) >DroSec_CAF1 185692 94 + 1 -CGCACCUAGCACCAGCUCCU---------------CGGAUCCA--------GAAAAUCUGCGCAUAAGCAGCUUGACUCUUGCCAGAUUGGAUGCGAUGGAGGUG--GGCAAUCCUGCC -.(((....((.(((.(((((---------------((.(((((--------....(((((.(((..(((.....).))..))))))))))))).))).)))).))--))).....))). ( -37.90) >DroSim_CAF1 180743 98 + 1 -CGCACCUAGCACCAGCUCCU---------------CGGAUCCAAA----GCGAAAAUCUGCGCAUAAGCAGCUUGACUCUUGCCAGAUUGGAUGCGAUGGAGGUG--GGCAAUCCUGCC -.(((....((.(((.(((((---------------((.(((((..----......(((((.(((..(((.....).))..))))))))))))).))).)))).))--))).....))). ( -36.80) >DroEre_CAF1 175073 102 + 1 -CACGCGCAGCACCAGCUCCC---------------CGGAUCCAAAGACUGCGAAAAUCUGCGCAUAAGCAGCUCGACUCUGGCCAGAUUGGAUGCGAUGGAGGUG--GGCAAUCCUGCC -.....((((..(((.((((.---------------((.((((((......(((....((((......)))).)))..(((....))))))))).))..)))).))--)......)))). ( -33.50) >DroYak_CAF1 178694 119 + 1 -CACUCGCAGCACCAACCCCCCCUUCCCCCUCCCCCUGUAUCCAAAAUCUGCGAAAAUCUGCGCAUAAGCAGCUUGACUCUUGCCAGAUUGGAUGCAAUGGAGGUGGAGGCAAUCCUACC -.....................(((((.(((((...((((((((..((((((((.((.((((......)))).)).....))).)))))))))))))..))))).))))).......... ( -37.30) >consensus _CACACCCAGCACCAGCUCCU_______________CGGAUCCAAA____GCGAAAAUCUGCGCAUAAGCAGCUUGACUCUUGCCAGAUUGGAUGCGAUGGAGGUG__GGCAAUCCUGCC .........((..((.((((................((.(((((............(((((.(((..(((.....).))..))))))))))))).))..)))).))...))......... (-21.43 = -21.35 + -0.08)

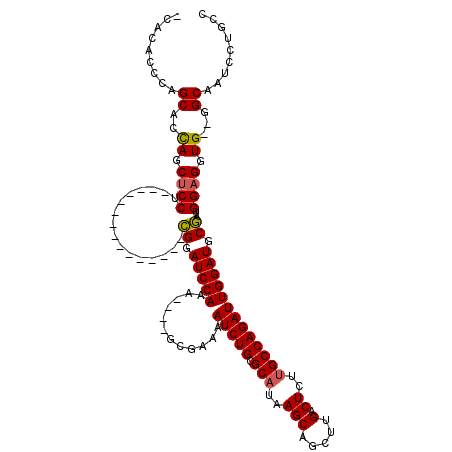

| Location | 11,453,202 – 11,453,311 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.91 |
| Mean single sequence MFE | -39.98 |
| Consensus MFE | -27.42 |
| Energy contribution | -27.22 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.07 |
| Mean z-score | -3.28 |
| Structure conservation index | 0.69 |
| SVM decision value | 1.61 |
| SVM RNA-class probability | 0.967285 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11453202 109 + 27905053 UCCAAA----GCGAAAAUCUGCGCAUAAGCAGCUUGACUCUUGCCAGAUUGGAUGCGAUGGAGGUG--GGCAAUCCUGCCCAGCCCAACC-----CACUGCAUCCACCGCACUUCACUCG ......----(((..((((((.(((..(((.....).))..)))))))))(((((((.(((.((((--((((....)))))).))....)-----)).)))))))..))).......... ( -41.90) >DroSec_CAF1 185716 105 + 1 UCCA--------GAAAAUCUGCGCAUAAGCAGCUUGACUCUUGCCAGAUUGGAUGCGAUGGAGGUG--GGCAAUCCUGCCCAGCCCAACC-----CACUGCAUCCACCGCACUUCACUCG ....--------(((((((((.(((..(((.....).))..)))))))))(((((((.(((.((((--((((....)))))).))....)-----)).))))))).......)))..... ( -38.40) >DroSim_CAF1 180767 109 + 1 UCCAAA----GCGAAAAUCUGCGCAUAAGCAGCUUGACUCUUGCCAGAUUGGAUGCGAUGGAGGUG--GGCAAUCCUGCCCAGCCCAACC-----CACUGCAUCCACCGCACUUCACUCG ......----(((..((((((.(((..(((.....).))..)))))))))(((((((.(((.((((--((((....)))))).))....)-----)).)))))))..))).......... ( -41.90) >DroEre_CAF1 175097 101 + 1 UCCAAAGACUGCGAAAAUCUGCGCAUAAGCAGCUCGACUCUGGCCAGAUUGGAUGCGAUGGAGGUG--GGCAAUCCUGCCCAGCCCGGCC-----CACUGCAUCCACC------------ .(((.((....(((....((((......)))).))).)).)))......((((((((.(((.((((--((((....))))))..))...)-----)).))))))))..------------ ( -36.50) >DroYak_CAF1 178733 120 + 1 UCCAAAAUCUGCGAAAAUCUGCGCAUAAGCAGCUUGACUCUUGCCAGAUUGGAUGCAAUGGAGGUGGAGGCAAUCCUACCCAGCCCAGCCCAAUCCACUGCAUCCACCACACUUCGCUCG ..........(((((((((((.(((..(((.....).))..)))))))))(((((((.(((((((.(.(((...........)))).)))...)))).))))))).......)))))... ( -41.20) >consensus UCCAAA____GCGAAAAUCUGCGCAUAAGCAGCUUGACUCUUGCCAGAUUGGAUGCGAUGGAGGUG__GGCAAUCCUGCCCAGCCCAACC_____CACUGCAUCCACCGCACUUCACUCG ...............((((((.(((..(((.....).))..)))))))))(((((((.((..(((...(((...........)))..))).....)).)))))))............... (-27.42 = -27.22 + -0.20)


| Location | 11,453,202 – 11,453,311 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.91 |
| Mean single sequence MFE | -45.52 |
| Consensus MFE | -28.76 |
| Energy contribution | -29.52 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.51 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.659647 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11453202 109 - 27905053 CGAGUGAAGUGCGGUGGAUGCAGUG-----GGUUGGGCUGGGCAGGAUUGCC--CACCUCCAUCGCAUCCAAUCUGGCAAGAGUCAAGCUGCUUAUGCGCAGAUUUUCGC----UUUGGA .((((((((.....(((((((.(((-----(...(((.(((((......)))--))))))))).)))))))((((((((.((((......)))).))).)))))))))))----)).... ( -47.40) >DroSec_CAF1 185716 105 - 1 CGAGUGAAGUGCGGUGGAUGCAGUG-----GGUUGGGCUGGGCAGGAUUGCC--CACCUCCAUCGCAUCCAAUCUGGCAAGAGUCAAGCUGCUUAUGCGCAGAUUUUC--------UGGA ...........(((.((((((.(((-----(...(((.(((((......)))--))))))))).))))))(((((((((.((((......)))).))).))))))..)--------)).. ( -41.70) >DroSim_CAF1 180767 109 - 1 CGAGUGAAGUGCGGUGGAUGCAGUG-----GGUUGGGCUGGGCAGGAUUGCC--CACCUCCAUCGCAUCCAAUCUGGCAAGAGUCAAGCUGCUUAUGCGCAGAUUUUCGC----UUUGGA .((((((((.....(((((((.(((-----(...(((.(((((......)))--))))))))).)))))))((((((((.((((......)))).))).)))))))))))----)).... ( -47.40) >DroEre_CAF1 175097 101 - 1 ------------GGUGGAUGCAGUG-----GGCCGGGCUGGGCAGGAUUGCC--CACCUCCAUCGCAUCCAAUCUGGCCAGAGUCGAGCUGCUUAUGCGCAGAUUUUCGCAGUCUUUGGA ------------..(((((((.(((-----(...(((.(((((......)))--))))))))).)))))))......((((((.(((.((((......))))....)))....)))))). ( -42.80) >DroYak_CAF1 178733 120 - 1 CGAGCGAAGUGUGGUGGAUGCAGUGGAUUGGGCUGGGCUGGGUAGGAUUGCCUCCACCUCCAUUGCAUCCAAUCUGGCAAGAGUCAAGCUGCUUAUGCGCAGAUUUUCGCAGAUUUUGGA ...((((((.....(((((((((((((...((.(((...(((((....)))))))))))))))))))))))((((((((.((((......)))).))).))))))))))).......... ( -48.30) >consensus CGAGUGAAGUGCGGUGGAUGCAGUG_____GGUUGGGCUGGGCAGGAUUGCC__CACCUCCAUCGCAUCCAAUCUGGCAAGAGUCAAGCUGCUUAUGCGCAGAUUUUCGC____UUUGGA .........(((((((((..((((............))))(((......)))......)))))))))(((((....((.((((((..((.((....)))).)))))).)).....))))) (-28.76 = -29.52 + 0.76)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:26:23 2006