
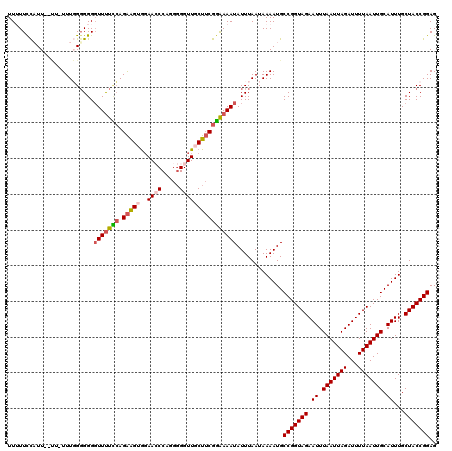
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 11,443,917 – 11,444,034 |
| Length | 117 |
| Max. P | 0.994349 |

| Location | 11,443,917 – 11,444,034 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.33 |
| Mean single sequence MFE | -31.42 |
| Consensus MFE | -21.88 |
| Energy contribution | -22.60 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.631043 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11443917 117 + 27905053 UUUUUCCAUU--UU-UUUGGUGGGCUUUUCCAGAAGUAGAACCCAGGGGGUUGCUUCGGAAAAUAUUUAAUAAAAUGCCGGUAGAAUUUAAUUAGAUUUUAAUUGCAUUUGCUACCGGAG ..........--..-((((((((..((((((.(((((((..((...))..))))))))))))).........((((((...((((((((....))))))))...)))))).)))))))). ( -33.90) >DroSec_CAF1 183873 118 + 1 UUUUUCCAUU--UUUUUUGGGGGGGUUUUCCAGAAGUGGAACCCAGGGGGUUGCUUCGGAAAAUAUUUAAUAAAAUGCCGGUAGAAUUUAAUUAGAUUUUAAUUGCAUUUGCUACCGGAG .((..(((..--.....)))..))(((((((.(((((..(((((...))))))))))))))))).............(((((((.((.(((((((...))))))).))...))))))).. ( -34.80) >DroSim_CAF1 178457 117 + 1 UUUUUCCAUU--UU-UUUGGGGGGGUUUUCCAGAAGUGGAACCCAGGGGGUUGCUUCGGAAAAUAUUUAAUAAAAUGCCGGUAGAAUUUAAUUAGAUUUUAAUUGCAUUUGCUACCGGAG .((..(((..--..-..)))..))(((((((.(((((..(((((...))))))))))))))))).............(((((((.((.(((((((...))))))).))...))))))).. ( -35.30) >DroEre_CAF1 173281 115 + 1 UUGUUUUUUU--A--UUUUGGGGGGUUCUGCAGAGGAGGAAGCCAGG-GGUUGCUUCGCGAAAUAUUUAAUAAAAUGCCGGUAGAAUUUAAUUAGAUUUUAAUUGCAUUUGCUACCGGAG (((((....(--(--(((((.(((((.((.(...((......)).).-))..))))).)))))))...)))))....(((((((.((.(((((((...))))))).))...))))))).. ( -27.20) >DroYak_CAF1 176831 118 + 1 UUAUUUUUUUCAC--UUUUGGGGGGUUUUUGAGGAGAGCAAGCCAGGGGGUUGCUACGAAAAAUAUUUAAUAAAAUGCCGGUAGAAUUUAAUUAGAUUUUAAUUGCAUUUGCUACCGGAG .....((((((.(--((((.(((....))).)))))(((((.((...)).)))))..))))))..............(((((((.((.(((((((...))))))).))...))))))).. ( -25.90) >consensus UUUUUCCAUU__UU_UUUGGGGGGGUUUUCCAGAAGUGGAACCCAGGGGGUUGCUUCGGAAAAUAUUUAAUAAAAUGCCGGUAGAAUUUAAUUAGAUUUUAAUUGCAUUUGCUACCGGAG ........................(((((((.(((((..((((.....)))))))))))))))).............(((((((.((.(((((((...))))))).))...))))))).. (-21.88 = -22.60 + 0.72)


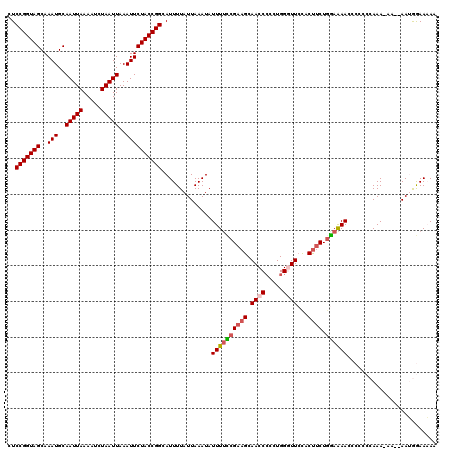
| Location | 11,443,917 – 11,444,034 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.33 |
| Mean single sequence MFE | -23.08 |
| Consensus MFE | -17.74 |
| Energy contribution | -18.22 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.10 |
| Mean z-score | -3.28 |
| Structure conservation index | 0.77 |
| SVM decision value | 2.47 |
| SVM RNA-class probability | 0.994349 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11443917 117 - 27905053 CUCCGGUAGCAAAUGCAAUUAAAAUCUAAUUAAAUUCUACCGGCAUUUUAUUAAAUAUUUUCCGAAGCAACCCCCUGGGUUCUACUUCUGGAAAAGCCCACCAAA-AA--AAUGGAAAAA .((((((((..(((..(((((.....)))))..))))))))(((((((....)))).((((((((((.(((((...)))))...)))).))))))))).......-..--...))).... ( -27.50) >DroSec_CAF1 183873 118 - 1 CUCCGGUAGCAAAUGCAAUUAAAAUCUAAUUAAAUUCUACCGGCAUUUUAUUAAAUAUUUUCCGAAGCAACCCCCUGGGUUCCACUUCUGGAAAACCCCCCCAAAAAA--AAUGGAAAAA ..(((((((..(((..(((((.....)))))..))))))))))..............((((((((((.(((((...)))))...)))).)))))).....(((.....--..)))..... ( -25.70) >DroSim_CAF1 178457 117 - 1 CUCCGGUAGCAAAUGCAAUUAAAAUCUAAUUAAAUUCUACCGGCAUUUUAUUAAAUAUUUUCCGAAGCAACCCCCUGGGUUCCACUUCUGGAAAACCCCCCCAAA-AA--AAUGGAAAAA ..(((((((..(((..(((((.....)))))..))))))))))..............((((((((((.(((((...)))))...)))).)))))).....(((..-..--..)))..... ( -26.20) >DroEre_CAF1 173281 115 - 1 CUCCGGUAGCAAAUGCAAUUAAAAUCUAAUUAAAUUCUACCGGCAUUUUAUUAAAUAUUUCGCGAAGCAACC-CCUGGCUUCCUCCUCUGCAGAACCCCCCAAAA--U--AAAAAAACAA ..(((((((..(((..(((((.....)))))..))))))))))..(((((((...........(((((....-....)))))....((....)).........))--)--))))...... ( -18.30) >DroYak_CAF1 176831 118 - 1 CUCCGGUAGCAAAUGCAAUUAAAAUCUAAUUAAAUUCUACCGGCAUUUUAUUAAAUAUUUUUCGUAGCAACCCCCUGGCUUGCUCUCCUCAAAAACCCCCCAAAA--GUGAAAAAAAUAA ..(((((((..(((..(((((.....)))))..))))))))))..............(((((((.(((((((....)).)))))).........((.........--))))))))..... ( -17.70) >consensus CUCCGGUAGCAAAUGCAAUUAAAAUCUAAUUAAAUUCUACCGGCAUUUUAUUAAAUAUUUUCCGAAGCAACCCCCUGGGUUCCACUUCUGGAAAACCCCCCCAAA_AA__AAUGGAAAAA ..(((((((..(((..(((((.....)))))..))))))))))..............((((((((((.((((.....))))...)))).))))))......................... (-17.74 = -18.22 + 0.48)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:26:17 2006