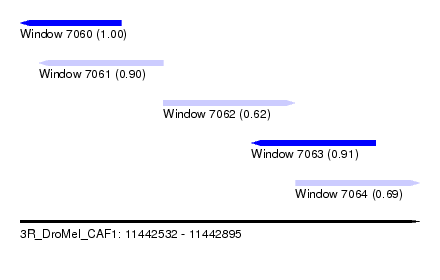
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 11,442,532 – 11,442,895 |
| Length | 363 |
| Max. P | 0.996887 |

| Location | 11,442,532 – 11,442,624 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.06 |
| Mean single sequence MFE | -35.28 |
| Consensus MFE | -22.50 |
| Energy contribution | -22.86 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.12 |
| Mean z-score | -3.09 |
| Structure conservation index | 0.64 |
| SVM decision value | 2.76 |
| SVM RNA-class probability | 0.996887 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11442532 92 - 27905053 CUUUUUUUUGGCUGCCGCGCAUUGGUGGCAACACCAAAUGGGCGUGGUAAUGAGC----UC-CAUUGAUGAUGAUCGUGAUGGAGGGUGAGGAAGUC----------------------- ...((((((.((((((((((.((((((....))))))....)))))))......(----((-((((.(((.....))))))))))))).))))))..----------------------- ( -38.40) >DroSec_CAF1 182487 88 - 1 CUCU-UUUUGGCUGCCGCGCAUUGGUGGCAACACCAAAUGGGCGUGGUAAUGAGC----UC-CAUUAA---UGAUCGUGAUGCAGGGAGAGGAAGUC----------------------- ((((-(((((((((((((((.((((((....))))))....)))))))....)))----..-(((((.---......))))))))))))))......----------------------- ( -35.00) >DroSim_CAF1 177058 112 - 1 CUCU-UUUUGGCUGCCGCGCAUUGGUGGCAACACCAAAUGGGCGUGGUAAUGAGC----UCCCAUUGA---UGAUCGUGAUGCAGGGAGAGGAAGGAAUNNNNNNNNNNNNNNNNNNNNN .(((-((((...((((((((.((((((....))))))....)))))))).....(----((((..(.(---(....)).)....))))).)))))))....................... ( -34.80) >DroEre_CAF1 171979 92 - 1 CUUU-UUUUGGCUACCGCUCAUUGGUGGCAACACCAAAGGGGCGUGGUAAUGAGAUCGGUG-CAUUGA---UGAUCGUGUUGGAGGCAGAGGAAGUC----------------------- ((((-(((((.((.((((((.((((((....))))))..))))..........(((((...-......---))))).....)))).)))))))))..----------------------- ( -33.40) >DroYak_CAF1 175407 88 - 1 CUUU-UUUUGGCUACCGCUCAUUGGUGGCAACACCAAAUGGGCGUGGUAAUGAGG----UC-CACUGA---UGAUCGUGAUGGGGGCAGAGGAAGUC----------------------- ((((-(((((.((.(((((((((((((....)))).)))))))((((........----.)-)))...---..........)))).)))))))))..----------------------- ( -34.80) >consensus CUUU_UUUUGGCUGCCGCGCAUUGGUGGCAACACCAAAUGGGCGUGGUAAUGAGC____UC_CAUUGA___UGAUCGUGAUGGAGGGAGAGGAAGUC_______________________ ((((.((((...((((((((.((((((....))))))....))))))))((((.....................))))....)))).))))............................. (-22.50 = -22.86 + 0.36)

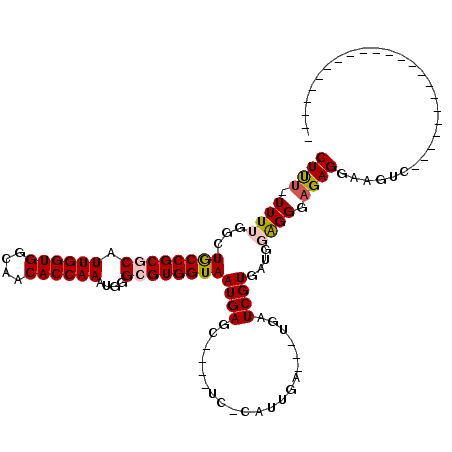
| Location | 11,442,549 – 11,442,662 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.18 |
| Mean single sequence MFE | -32.90 |
| Consensus MFE | -24.58 |
| Energy contribution | -24.78 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.75 |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.898741 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11442549 113 - 27905053 CGCGUUUUGUAG--CGUAUUUUCGUAUUUUGUACUUUAGUCUUUUUUUUGGCUGCCGCGCAUUGGUGGCAACACCAAAUGGGCGUGGUAAUGAGC----UC-CAUUGAUGAUGAUCGUGA (((....((.((--(((((...........))))...((((........))))(((((((.((((((....))))))....))))))).....))----).-))..(((....)))))). ( -32.80) >DroSec_CAF1 182504 109 - 1 CGCAUUUUGUAG--CGUAUUUUCGUAUUUUGUACUUUAGUCUCU-UUUUGGCUGCCGCGCAUUGGUGGCAACACCAAAUGGGCGUGGUAAUGAGC----UC-CAUUAA---UGAUCGUGA ..((((.((.((--(((((...........))))...((((...-....))))(((((((.((((((....))))))....))))))).....))----).-))..))---))....... ( -32.20) >DroSim_CAF1 177098 110 - 1 CGCAUUUUGUAG--CGUAUUUUCGUAUUUUGUACUUUAGUCUCU-UUUUGGCUGCCGCGCAUUGGUGGCAACACCAAAUGGGCGUGGUAAUGAGC----UCCCAUUGA---UGAUCGUGA ..((((.((.((--(((((...........))))...((((...-....))))(((((((.((((((....))))))....))))))).....))----)..))..))---))....... ( -30.70) >DroEre_CAF1 171996 101 - 1 CGCAUUUUGUCGGCCGCAU--------------CUUCGGUCUUU-UUUUGGCUACCGCUCAUUGGUGGCAACACCAAAGGGGCGUGGUAAUGAGAUCGGUG-CAUUGA---UGAUCGUGU .((((.(..((((..((((--------------...(((((((.-.....(((((.((((.((((((....))))))..)))))))))...))))))))))-).))))---..)..)))) ( -38.60) >DroYak_CAF1 175424 89 - 1 CGCAUUUUGUA----------------------CUUCAAUCUUU-UUUUGGCUACCGCUCAUUGGUGGCAACACCAAAUGGGCGUGGUAAUGAGG----UC-CACUGA---UGAUCGUGA ..((((.((.(----------------------(((((..(...-....)(((((.(((((((((((....)))).))))))))))))..)))))----).-))..))---))....... ( -30.20) >consensus CGCAUUUUGUAG__CGUAUUUUCGUAUUUUGUACUUUAGUCUUU_UUUUGGCUGCCGCGCAUUGGUGGCAACACCAAAUGGGCGUGGUAAUGAGC____UC_CAUUGA___UGAUCGUGA (((..................................((((........))))(((((((.((((((....))))))....)))))))............................))). (-24.58 = -24.78 + 0.20)

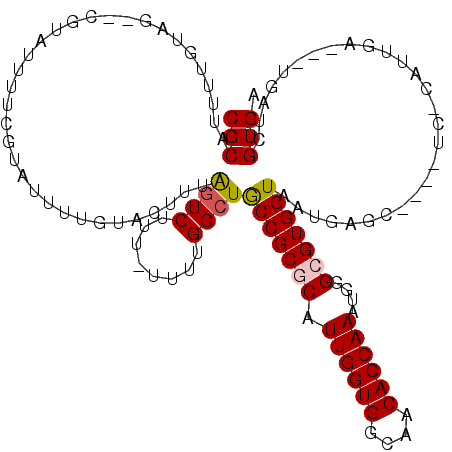

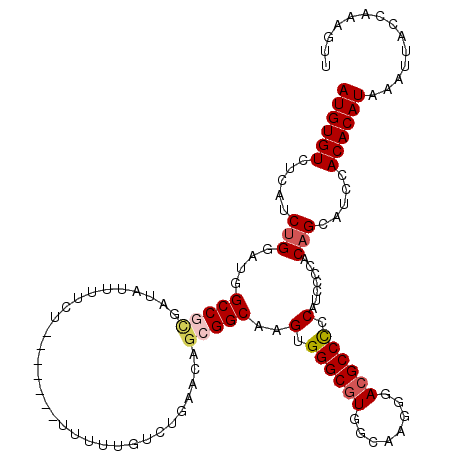
| Location | 11,442,662 – 11,442,782 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.50 |
| Mean single sequence MFE | -37.98 |
| Consensus MFE | -33.58 |
| Energy contribution | -34.22 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.617826 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11442662 120 + 27905053 AGGAAAGCGGCGGCAUUCAUCCGUGGCACCUUCACUGCUUCCCAUGCCUCAUGAAGUGAACUCAACUCUUCCUGGGGGCAGGCACGCCCACUUUUCCAAGUGAUCGCCACCGUUUCAGCC .((.((((((.(((.......(((((((.......)))...((.((((((..((((.(.......).))))...))))))))))))..(((((....)))))...))).))))))...)) ( -38.30) >DroSec_CAF1 182613 120 + 1 AGGAAAGCGGCGGCAUCCAUCCGUGGCACCUUCACUGCUUCCCAUGCCUCAUGAAGUGAACUCAACUCUUCCUGGGGGCAGGCACGCCCACUUUUCCAAGUGAUCGCCACCGUUUCAGCC .((.((((((.(((.......(((((((.......)))...((.((((((..((((.(.......).))))...))))))))))))..(((((....)))))...))).))))))...)) ( -38.30) >DroSim_CAF1 177208 120 + 1 AGGAAAGCGGCGGCAUCCAUCCGUGGCACCUUCACUGCUUCCCAUGCCUCAUGAAGUGAACUCAACUCUUCCUGGGGGCAGGCACGCCCACUUUUCCAAGUGAUCGCCACCGUUUCAGCC .((.((((((.(((.......(((((((.......)))...((.((((((..((((.(.......).))))...))))))))))))..(((((....)))))...))).))))))...)) ( -38.30) >DroEre_CAF1 172097 120 + 1 AGGAAAGCGGCGGCAUCCAUCCGUGGCACCUUCACUUCUCACCAUGCCUCAUGAAGUGAACUCAACUCUUCCUGGGGGCAGGCACGCCCCCUUUUCCAAGUGAUCGCCAACGUUUCAGCC .((.(((((((((.......))))(((...((((((((..............)))))))).(((.........((((((......)))))).........)))..)))..)))))...)) ( -37.61) >DroYak_CAF1 175513 120 + 1 AGGAAAGCGGCGGCAUCCAUCCGUGGCACCUUCACUUCUUCCCAUGCCUCAUGGAGUGAACUCAACUCUUCCUGGGGGCAGGCACGCCCACUUUUCCAAGUGAUCGCCACCGUUUCAGCC .((.((((((.(((..(((....))).....((((((.......((((((..(((((.......))))).....))))))((....)).........))))))..))).))))))...)) ( -37.40) >consensus AGGAAAGCGGCGGCAUCCAUCCGUGGCACCUUCACUGCUUCCCAUGCCUCAUGAAGUGAACUCAACUCUUCCUGGGGGCAGGCACGCCCACUUUUCCAAGUGAUCGCCACCGUUUCAGCC .((.((((((.(((.......(((((((.......)))...((.((((((..((((.(.......).))))...))))))))))))..(((((....)))))...))).))))))...)) (-33.58 = -34.22 + 0.64)

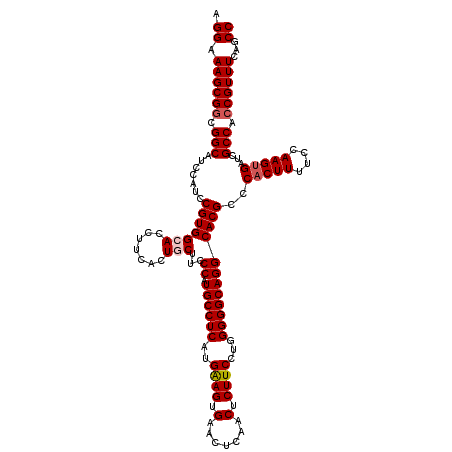
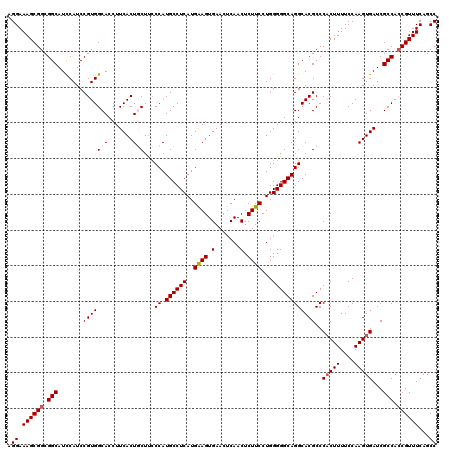
| Location | 11,442,742 – 11,442,855 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.86 |
| Mean single sequence MFE | -38.37 |
| Consensus MFE | -31.99 |
| Energy contribution | -33.27 |
| Covariance contribution | 1.28 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.04 |
| SVM RNA-class probability | 0.905922 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11442742 113 - 27905053 CUUCCCUUGCCACGCCCACUUGCCGCUGCUCAGACAAAAA-------AAAAAAAAGAAAGGCCAUCCAGAUGAGACACAUGGCUGAAACGGUGGCGAUCACUUGGAAAAGUGGGCGUGCC ..........((((((((((((((.((.............-------.......))...)))..(((((.(((....(((.(......).)))....))).))))).))))))))))).. ( -35.05) >DroSec_CAF1 182693 118 - 1 CGUUCCUUGCCACGCCCACUUGCCGCUGCUCAGACAAAAA--AAAGAAGAAAAUAUCGCGGCCAUCCAGAUGAGACACAUGGCUGAAACGGUGGCGAUCACUUGGAAAAGUGGGCGUGCC ..........((((((((((((((((((......))....--......((.....)))))))..(((((.(((....(((.(......).)))....))).))))).))))))))))).. ( -39.20) >DroSim_CAF1 177288 113 - 1 CGUUCCUUGCCACGCCCACUUGCCGCUGUACAGACAAAAA-------AGAAAAUAUCGCGGCCAUCCAGAUGAGACACAUGGCUGAAACGGUGGCGAUCACUUGGAAAAGUGGGCGUGCC ..........(((((((((((((((((((....)))....-------.((.....)))))))..(((((.(((....(((.(......).)))....))).))))).))))))))))).. ( -40.70) >DroEre_CAF1 172177 92 - 1 CGUCUCUUGCCACGCCCACUUGCCGCUGUUC----------------------------GGCUGUUCGGAGGAGACACAUGGCUGAAACGUUGGCGAUCACUUGGAAAAGGGGGCGUGCC ..........(((((((..((((((.(((((----------------------------((((((..(..(....).)))))))).)))).))))))...(((....))).))))))).. ( -34.80) >DroYak_CAF1 175593 120 - 1 CGUCUCUUGCCACGCCCACUUGCCGCUGUUCACACAAAAAAAAAAGAAGAAAAUAUCGCGGCCAGCCAGAUGAGACACAUGGCUGAAACGGUGGCGAUCACUUGGAAAAGUGGGCGUGCC ..........((((((((((((((((((((..................((.....)).....((((((..((...))..)))))).)))))))))..((.....)).))))))))))).. ( -42.10) >consensus CGUCCCUUGCCACGCCCACUUGCCGCUGUUCAGACAAAAA_______AGAAAAUAUCGCGGCCAUCCAGAUGAGACACAUGGCUGAAACGGUGGCGAUCACUUGGAAAAGUGGGCGUGCC ..........((((((((((((((((((((............................(((((((.............))))))).)))))))))..((.....)).))))))))))).. (-31.99 = -33.27 + 1.28)

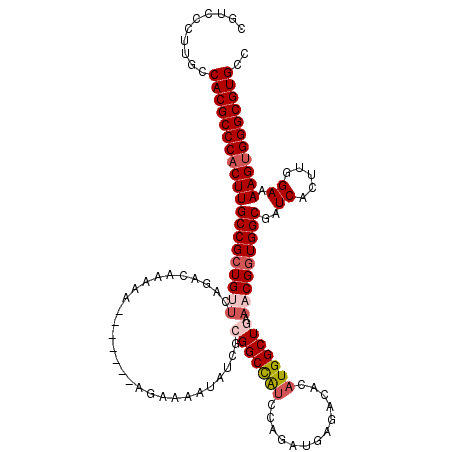
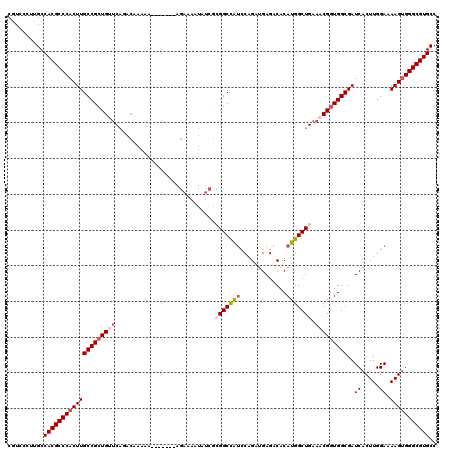
| Location | 11,442,782 – 11,442,895 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.99 |
| Mean single sequence MFE | -32.41 |
| Consensus MFE | -22.12 |
| Energy contribution | -22.84 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.688620 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11442782 113 + 27905053 AUGUGUCUCAUCUGGAUGGCCUUUCUUUUUUUU-------UUUUUGUCUGAGCAGCGGCAAGUGGGCGUGGCAAGGGAAGCCUCCAUCCCCACAGCAUCCACACAUAAAUUACCAAAGUU ((((((.......((((((((.(((((......-------.........))).)).)))..(((((.((((..(((....))))))).)))))..))))))))))).............. ( -32.27) >DroSec_CAF1 182733 118 + 1 AUGUGUCUCAUCUGGAUGGCCGCGAUAUUUUCUUCUUU--UUUUUGUCUGAGCAGCGGCAAGUGGGCGUGGCAAGGAACGCCCCCAUCCCCACAGCAUCCACACAUAAAUUACCAAAGUU ((((((.......((((((((((((.....))......--....(((....))))))))..(((((.((((...((....)).)))).)))))..))))))))))).............. ( -36.01) >DroSim_CAF1 177328 113 + 1 AUGUGUCUCAUCUGGAUGGCCGCGAUAUUUUCU-------UUUUUGUCUGUACAGCGGCAAGUGGGCGUGGCAAGGAACGCCCCCAUCCACACAGCAUCCACACAUAAAUUACCAAAGUU ((((((......(((((((((((..(((...(.-------.....)...)))..)))))....((((((........)))))).))))))..........)))))).............. ( -34.09) >DroEre_CAF1 172217 90 + 1 AUGUGUCUCCUCCGAACAGCC----------------------------GAACAGCGGCAAGUGGGCGUGGCAAGAGACGCCC-CAUC-CCACAGCAUCCACACAUAAAUAACCAAACUU ((((((............(((----------------------------(.....))))..(((((.(.(((.......))).-)..)-)))).......)))))).............. ( -24.30) >DroYak_CAF1 175633 120 + 1 AUGUGUCUCAUCUGGCUGGCCGCGAUAUUUUCUUCUUUUUUUUUUGUGUGAACAGCGGCAAGUGGGCGUGGCAAGAGACGCCCCCAUCCCCACAGCAUCCACACAUAAAUAACGAAAGUU ((((((........(((((((((((.....))............(((....))))))))..(.((((((........)))))).).......))))....))))))....(((....))) ( -35.40) >consensus AUGUGUCUCAUCUGGAUGGCCGCGAUAUUUUCU_______UUUUUGUCUGAACAGCGGCAAGUGGGCGUGGCAAGGGACGCCCCCAUCCCCACAGCAUCCACACAUAAAUUACCAAAGUU ((((((.....(((....(((((...............................)))))..(.((((((........)))))).).......))).....)))))).............. (-22.12 = -22.84 + 0.72)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:26:10 2006