
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 11,441,347 – 11,441,484 |
| Length | 137 |
| Max. P | 0.996563 |

| Location | 11,441,347 – 11,441,444 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.14 |
| Mean single sequence MFE | -33.66 |
| Consensus MFE | -28.10 |
| Energy contribution | -28.14 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.75 |
| Structure conservation index | 0.83 |
| SVM decision value | 2.72 |
| SVM RNA-class probability | 0.996563 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11441347 97 + 27905053 ----UAAUUGUUAUG-------------------AUUUAAGCCCAUAUGCUUUGCCGAUGCAGGAGUAUCCCCCAAAAGGGAUAUCGAUAAAUGCAGGCAAAUAUUGCAAUGGCGAGGCC ----...........-------------------..............(((((((((.(((((((.((((((......))))))))......((....))....))))).))))))))). ( -28.10) >DroSec_CAF1 181286 116 + 1 ----UAAUUGUUAUGUUGAUUUAAAUAAACUAUUAUUUAAGCCGAUAUGCCUUGCCGAUGCAUGGGUAUCCCCCACAAGGGAUAUCGGUAAAUGCUGGCAAAUAUUGCAAUGGCGAGGCC ----........((((((..((((((((....))))))))..))))))(((((((((.((((..((((((((......))))))))(((....))).........)))).))))))))). ( -41.80) >DroSim_CAF1 175495 116 + 1 ----UAAUUGUUAUGUUGAUUUAAAUAAACUAUUAUUUAAGCCCAUAUGCCUUGCCGAUGCAUGGGUAUCCCCCACAAGGGAUAUCGGUAAAUGCUGGCAAAUAUUGCAAUGGCGAGGCC ----........((((.(..((((((((....))))))))..).))))(((((((((.((((..((((((((......))))))))(((....))).........)))).))))))))). ( -38.00) >DroEre_CAF1 170787 111 + 1 GUUAUAACUAUUAUC-CCACUUAAACAAACU--GGU-UCAGCCCAUAUGCUUUGCCGAUGCAGGAGUAUCCC-----AGGGAUAUUGGUAAAUGCAGGCAAAUAUUGCAAUGGCGAGGCC ((....((((.((((-((.............--(((-..(((......)))..)))(((((....)))))..-----.)))))).))))....)).(((.....((((....)))).))) ( -30.80) >DroYak_CAF1 174201 119 + 1 ACCAUAACUAUUAUUUUCAUUUAAGUAAACUAUGAU-UCAACCCAUAUGCUUGGCCGAUGCAGUAGUAUGCCCCAGAAGGGAUAUUGGUAAAUGCACGCAAAUAUUGCAAUGGCGAGGCC ..((((....((((((......))))))..))))..-...........((((.((((.(((((((...((((((....))).((((....))))...)))..))))))).)))).)))). ( -29.60) >consensus ____UAAUUGUUAUGUUCAUUUAAAUAAACUAUGAUUUAAGCCCAUAUGCUUUGCCGAUGCAGGAGUAUCCCCCACAAGGGAUAUCGGUAAAUGCAGGCAAAUAUUGCAAUGGCGAGGCC ................................................(((((((((.((((..((((((((......))))))))......((....)).....)))).))))))))). (-28.10 = -28.14 + 0.04)

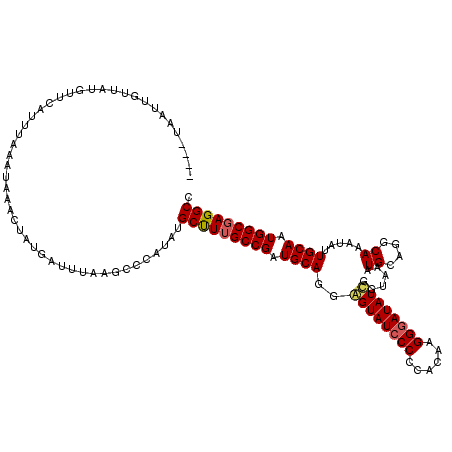
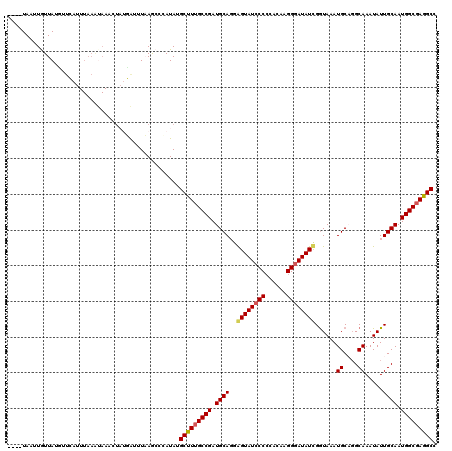
| Location | 11,441,347 – 11,441,444 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 120 |
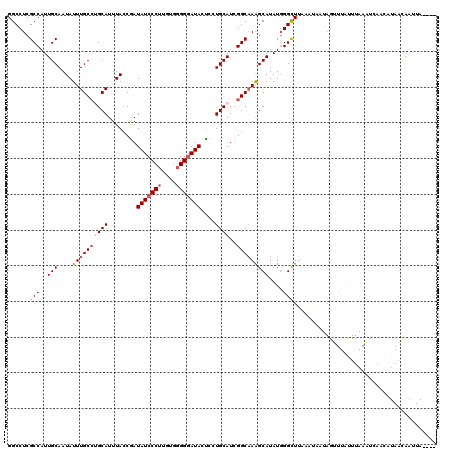
| Reading direction | reverse |
| Mean pairwise identity | 79.14 |
| Mean single sequence MFE | -27.52 |
| Consensus MFE | -19.48 |
| Energy contribution | -20.28 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.71 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.528536 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11441347 97 - 27905053 GGCCUCGCCAUUGCAAUAUUUGCCUGCAUUUAUCGAUAUCCCUUUUGGGGGAUACUCCUGCAUCGGCAAAGCAUAUGGGCUUAAAU-------------------CAUAACAAUUA---- ......(((..(((....((((((((((......(((((((((....))))))).)).))))..)))))))))....)))......-------------------...........---- ( -25.40) >DroSec_CAF1 181286 116 - 1 GGCCUCGCCAUUGCAAUAUUUGCCAGCAUUUACCGAUAUCCCUUGUGGGGGAUACCCAUGCAUCGGCAAGGCAUAUCGGCUUAAAUAAUAGUUUAUUUAAAUCAACAUAACAAUUA---- .((((.(((..((((.((..((....))..))....(((((((....)))))))....))))..))).)))).....(..((((((((....))))))))..).............---- ( -32.70) >DroSim_CAF1 175495 116 - 1 GGCCUCGCCAUUGCAAUAUUUGCCAGCAUUUACCGAUAUCCCUUGUGGGGGAUACCCAUGCAUCGGCAAGGCAUAUGGGCUUAAAUAAUAGUUUAUUUAAAUCAACAUAACAAUUA---- .((((.(((..((((.((..((....))..))....(((((((....)))))))....))))..))).)))).(((((..((((((((....))))))))..)..)))).......---- ( -33.00) >DroEre_CAF1 170787 111 - 1 GGCCUCGCCAUUGCAAUAUUUGCCUGCAUUUACCAAUAUCCCU-----GGGAUACUCCUGCAUCGGCAAAGCAUAUGGGCUGA-ACC--AGUUUGUUUAAGUGG-GAUAAUAGUUAUAAC (((...)))..((((.........))))........((((((.-----(((.....)))((....)).(((((..(((.....-.))--)...)))))....))-))))........... ( -23.20) >DroYak_CAF1 174201 119 - 1 GGCCUCGCCAUUGCAAUAUUUGCGUGCAUUUACCAAUAUCCCUUCUGGGGCAUACUACUGCAUCGGCCAAGCAUAUGGGUUGA-AUCAUAGUUUACUUAAAUGAAAAUAAUAGUUAUGGU ((((........(((.....)))(((((.......((..(((....)))..)).....))))).))))((((.(((((.....-.))))))))).......................... ( -23.30) >consensus GGCCUCGCCAUUGCAAUAUUUGCCUGCAUUUACCGAUAUCCCUUGUGGGGGAUACUCCUGCAUCGGCAAAGCAUAUGGGCUUAAAUAAUAGUUUAUUUAAAUCAACAUAACAAUUA____ ......(((..(((....((((((((((........(((((((....)))))))....))))..)))))))))....)))........................................ (-19.48 = -20.28 + 0.80)


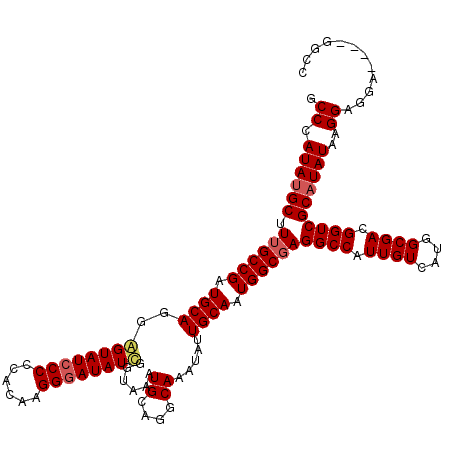
| Location | 11,441,364 – 11,441,484 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.96 |
| Mean single sequence MFE | -41.92 |
| Consensus MFE | -36.32 |
| Energy contribution | -36.80 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.61 |
| SVM RNA-class probability | 0.967395 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11441364 120 + 27905053 GCCCAUAUGCUUUGCCGAUGCAGGAGUAUCCCCCAAAAGGGAUAUCGAUAAAUGCAGGCAAAUAUUGCAAUGGCGAGGCCAUUGUCAUGGCGACGGUCGCAUAUAAGGAGGCGGCUGGCU (((.......((((((..((((...(((((((......))))))).......))))))))))....((((((((...))))))))...)))..(((((((..........)))))))... ( -42.80) >DroSec_CAF1 181322 111 + 1 GCCGAUAUGCCUUGCCGAUGCAUGGGUAUCCCCCACAAGGGAUAUCGGUAAAUGCUGGCAAAUAUUGCAAUGGCGAGGCCAUUGUCAUGGCGACGGUCGCAUAUAAGGAGG--------- .((((((.(((((((((.((((..((((((((......))))))))(((....))).........)))).)))))))))...))))...(((.....)))......))...--------- ( -42.80) >DroSim_CAF1 175531 120 + 1 GCCCAUAUGCCUUGCCGAUGCAUGGGUAUCCCCCACAAGGGAUAUCGGUAAAUGCUGGCAAAUAUUGCAAUGGCGAGGCCAUUGUCAUGGCGACGGUCGCAUAUAAGGAGGCGGCUGGCU (((((...(((((((((.((((..((((((((......))))))))(((....))).........)))).)))))))))...))....)))..(((((((..........)))))))... ( -47.70) >DroEre_CAF1 170823 111 + 1 GCCCAUAUGCUUUGCCGAUGCAGGAGUAUCCC-----AGGGAUAUUGGUAAAUGCAGGCAAAUAUUGCAAUGGCGAGGCCAUUGUCAUGGCGACGGUCGCAUAUAAGGAGCA----GGCC ((((((((((.((((((.(((((.(((((((.-----..)))))))......((....))....))))).))))))((((.((((....)))).))))))))))..)).)).----.... ( -38.20) >DroYak_CAF1 174240 116 + 1 ACCCAUAUGCUUGGCCGAUGCAGUAGUAUGCCCCAGAAGGGAUAUUGGUAAAUGCACGCAAAUAUUGCAAUGGCGAGGCCAUUGUCAUGGCGACGGUCGCCUAUAAGGAGGA----GGCC ........((((.((((.(((((((...((((((....))).((((....))))...)))..))))))).)))).))))....(((.....)))((((.(((......))).----)))) ( -38.10) >consensus GCCCAUAUGCUUUGCCGAUGCAGGAGUAUCCCCCACAAGGGAUAUCGGUAAAUGCAGGCAAAUAUUGCAAUGGCGAGGCCAUUGUCAUGGCGACGGUCGCAUAUAAGGAGGA____GGCC .((.((((((.((((((.((((..((((((((......))))))))......((....)).....)))).))))))((((.((((....)))).))))))))))..))............ (-36.32 = -36.80 + 0.48)


| Location | 11,441,364 – 11,441,484 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.96 |
| Mean single sequence MFE | -35.94 |
| Consensus MFE | -34.36 |
| Energy contribution | -35.16 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.96 |
| SVM decision value | 1.41 |
| SVM RNA-class probability | 0.951175 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11441364 120 - 27905053 AGCCAGCCGCCUCCUUAUAUGCGACCGUCGCCAUGACAAUGGCCUCGCCAUUGCAAUAUUUGCCUGCAUUUAUCGAUAUCCCUUUUGGGGGAUACUCCUGCAUCGGCAAAGCAUAUGGGC ........((((....((((((....(..(((((....)))))..)(((..(((((((..((....))..))).(((((((((....))))))).)).))))..)))...)))))))))) ( -39.30) >DroSec_CAF1 181322 111 - 1 ---------CCUCCUUAUAUGCGACCGUCGCCAUGACAAUGGCCUCGCCAUUGCAAUAUUUGCCAGCAUUUACCGAUAUCCCUUGUGGGGGAUACCCAUGCAUCGGCAAGGCAUAUCGGC ---------......((.((((...((..(((((....)))))..)).....(((.....)))..)))).))(((((((.(((((((((.....)))))((....)))))).))))))). ( -35.60) >DroSim_CAF1 175531 120 - 1 AGCCAGCCGCCUCCUUAUAUGCGACCGUCGCCAUGACAAUGGCCUCGCCAUUGCAAUAUUUGCCAGCAUUUACCGAUAUCCCUUGUGGGGGAUACCCAUGCAUCGGCAAGGCAUAUGGGC ..(((...(((........(((((.((..(((((....)))))..))...)))))....(((((.((((.....(.(((((((....))))))).).))))...))))))))...))).. ( -39.20) >DroEre_CAF1 170823 111 - 1 GGCC----UGCUCCUUAUAUGCGACCGUCGCCAUGACAAUGGCCUCGCCAUUGCAAUAUUUGCCUGCAUUUACCAAUAUCCCU-----GGGAUACUCCUGCAUCGGCAAAGCAUAUGGGC ....----.....(((((((((...........((.(((((((...)))))))))...((((((((((........(((((..-----.)))))....))))..))))))))))))))). ( -31.90) >DroYak_CAF1 174240 116 - 1 GGCC----UCCUCCUUAUAGGCGACCGUCGCCAUGACAAUGGCCUCGCCAUUGCAAUAUUUGCGUGCAUUUACCAAUAUCCCUUCUGGGGCAUACUACUGCAUCGGCCAAGCAUAUGGGU ((((----...........((((.....)))).((.(((((((...)))))))))........(((((.......((..(((....)))..)).....))))).))))............ ( -33.70) >consensus AGCC____GCCUCCUUAUAUGCGACCGUCGCCAUGACAAUGGCCUCGCCAUUGCAAUAUUUGCCUGCAUUUACCGAUAUCCCUUGUGGGGGAUACUCCUGCAUCGGCAAAGCAUAUGGGC .............(((((((((....(..(((((....)))))..)(((..((((.((..((....))..))....(((((((....)))))))....))))..)))...))))))))). (-34.36 = -35.16 + 0.80)


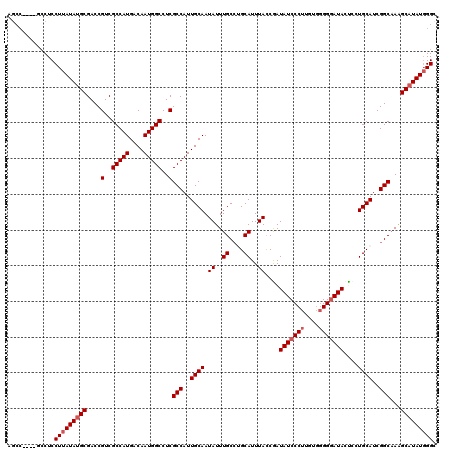
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:26:05 2006