
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
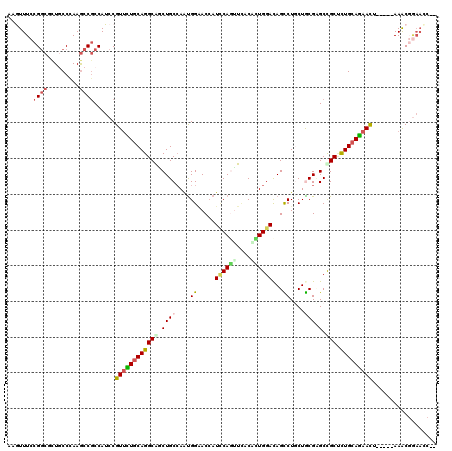
| Location | 1,624,011 – 1,624,124 |
| Length | 113 |
| Max. P | 0.946307 |

| Location | 1,624,011 – 1,624,124 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.83 |
| Mean single sequence MFE | -39.02 |
| Consensus MFE | -22.19 |
| Energy contribution | -23.00 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.43 |
| Mean z-score | -2.47 |
| Structure conservation index | 0.57 |
| SVM decision value | 1.30 |
| SVM RNA-class probability | 0.939196 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 1624011 113 + 27905053 GUGUUUCCGUCCCUGCCCAAGCCGCCAUCCGUUUUGCAGGCGGCUGCCAAUGGGACCAUACAGUUCACUCUGGACAGUCUUCUACGAGCCGCUCUGCAGAACU-----AAACGGAACC-- ....((((((....((....))........(((((((((((((((.(....(((((....(((......)))....)))))....))))))).))))))))).-----..))))))..-- ( -38.50) >DroGri_CAF1 3691 107 + 1 CUAUUUCCGGCGCUGCCCAAGCCGCCAUCCGUUCUGCAGGCAUCUGCGAAUGGAACAAUCCAAUUCACGCUGGACAGCCUGCUGCGUGCCGCUCUGCAGAAUU-----AAAC-------- ........((((((.....)).))))....(((((((((((.........((((....))))...(((((.((....))....)))))..)).))))))))).-----....-------- ( -36.10) >DroWil_CAF1 3666 113 + 1 AAGUUUCCGGCGUUGCCCAAGCCGCCAUCCGUUUUACAGGCAACUGCCAGUGGCACCAUUCAGUUUACAUUGGACAGCCUGCUGCGCGCCGCUUUGCAAAACU-----AAACGGAACC-- ........((((..((....)))))).(((((((....(((....)))((((((....((((((....))))))(((....)))...))))))..........-----)))))))...-- ( -33.40) >DroMoj_CAF1 3569 112 + 1 AAGUUUCCGGCGCUACCGAAACCCCCAUCCGUGCUGCAGGCGUCUGCCAACGGAACAAUCCAAUUCACAUUGGACAGCCUGCUGCGUGCUGCACUGCAGAACU-----AAGCCAAAU--- ........(((.....................((.((((((.((((....))))....((((((....))))))..)))))).))((.((((...)))).)).-----..)))....--- ( -33.30) >DroAna_CAF1 3447 118 + 1 ACGUUUCCGGCCCUGCCCAAGCCGCCAUCCGUUUUGCAGGCAGCUGCCAAUGGAACCAUCCAGCUAACGCUGGACAGUCUGCUACGAGCUGCUCUGCAGAACUAAACUAAACGGAACC-- ....(((((((...((....)).)))....(((((((((((((((......(((....((((((....))))))...)))......)))))).)))))))))..........))))..-- ( -46.30) >DroPer_CAF1 3221 115 + 1 AAAUUUCCGGCAUUACCGAAGCCGCCAUCCGUUCUGCAGGCAGCUGCCAAUGGAACCAUCCAGCUCACAUUGGAUAGUCUCCUGCGCGCUGCCCUGCAGAACU-----AAGCGGAACCAG ....(((((((.........)))((.....(((((((((((((((((....(((((.((((((......)))))).)).))).))).)))).)))))))))).-----..)))))).... ( -46.50) >consensus AAGUUUCCGGCGCUGCCCAAGCCGCCAUCCGUUCUGCAGGCAGCUGCCAAUGGAACCAUCCAGUUCACACUGGACAGCCUGCUGCGAGCCGCUCUGCAGAACU_____AAACGGAACC__ .......((((.........))))......((((((((((((.((((....((.....((((((....))))))...))....))).).))).))))))))).................. (-22.19 = -23.00 + 0.81)


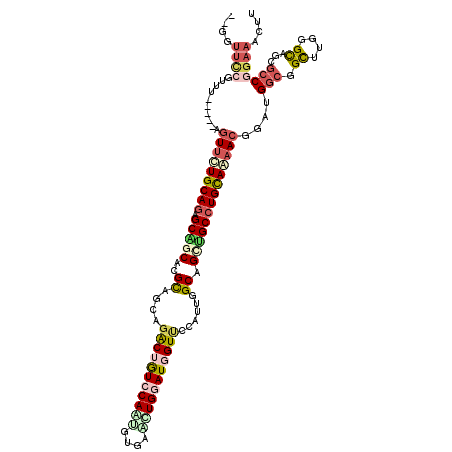
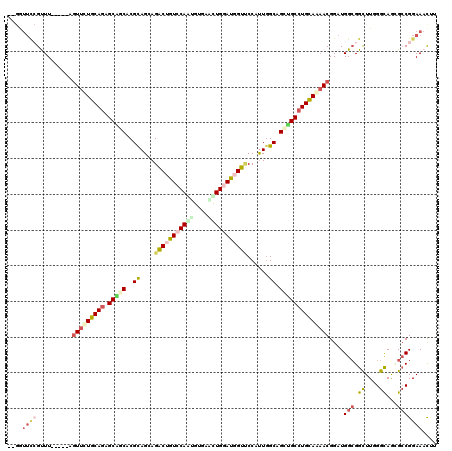
| Location | 1,624,011 – 1,624,124 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.83 |
| Mean single sequence MFE | -45.95 |
| Consensus MFE | -32.72 |
| Energy contribution | -34.48 |
| Covariance contribution | 1.76 |
| Combinations/Pair | 1.44 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.71 |
| SVM decision value | 1.36 |
| SVM RNA-class probability | 0.946307 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 1624011 113 - 27905053 --GGUUCCGUUU-----AGUUCUGCAGAGCGGCUCGUAGAAGACUGUCCAGAGUGAACUGUAUGGUCCCAUUGGCAGCCGCCUGCAAAACGGAUGGCGGCUUGGGCAGGGACGGAAACAC --.((((((((.-----.(((.(((((.((((((.((....((((((.(((......))).))))))......))))))))))))).)))....))).((....)).)))))(....).. ( -43.00) >DroGri_CAF1 3691 107 - 1 --------GUUU-----AAUUCUGCAGAGCGGCACGCAGCAGGCUGUCCAGCGUGAAUUGGAUUGUUCCAUUCGCAGAUGCCUGCAGAACGGAUGGCGGCUUGGGCAGCGCCGGAAAUAG --------....-----..((((((((.(((((..((((....))))...))((((((.(((....)))))))))...)))))))))))(((...((.((....)).)).)))....... ( -39.60) >DroWil_CAF1 3666 113 - 1 --GGUUCCGUUU-----AGUUUUGCAAAGCGGCGCGCAGCAGGCUGUCCAAUGUAAACUGAAUGGUGCCACUGGCAGUUGCCUGUAAAACGGAUGGCGGCUUGGGCAACGCCGGAAACUU --(((.((((((-----(((((.(((..(((((.(......))))))....)))))))))))))).))).(((((.((((((((.....((.....))...)))))))))))))...... ( -45.00) >DroMoj_CAF1 3569 112 - 1 ---AUUUGGCUU-----AGUUCUGCAGUGCAGCACGCAGCAGGCUGUCCAAUGUGAAUUGGAUUGUUCCGUUGGCAGACGCCUGCAGCACGGAUGGGGGUUUCGGUAGCGCCGGAAACUU ---.((((((..-----.((.((((...)))).))((.((((((((.((((((.((((......))))))))))))...)))))).((.((((.......)))))).))))))))..... ( -40.80) >DroAna_CAF1 3447 118 - 1 --GGUUCCGUUUAGUUUAGUUCUGCAGAGCAGCUCGUAGCAGACUGUCCAGCGUUAGCUGGAUGGUUCCAUUGGCAGCUGCCUGCAAAACGGAUGGCGGCUUGGGCAGGGCCGGAAACGU --(.((((.....(((..(((.(((((.((((((.((....(((((((((((....)))))))))))......))))))))))))).)))....)))(((((.....))))))))).).. ( -51.40) >DroPer_CAF1 3221 115 - 1 CUGGUUCCGCUU-----AGUUCUGCAGGGCAGCGCGCAGGAGACUAUCCAAUGUGAGCUGGAUGGUUCCAUUGGCAGCUGCCUGCAGAACGGAUGGCGGCUUCGGUAAUGCCGGAAAUUU (((((..((((.-----.((((((((((.((((((.((((..((((((((........))))))))..).))))).))))))))))))))....))))((....))...)))))...... ( -55.90) >consensus __GGUUCCGUUU_____AGUUCUGCAGAGCAGCACGCAGCAGACUGUCCAAUGUGAACUGGAUGGUUCCAUUGGCAGCUGCCUGCAAAACGGAUGGCGGCUUGGGCAGCGCCGGAAACUU ....((((..........(((((((((.(((((..((....(((((((((((....)))))))))))......)).))))))))))))))....(((.((....))...))))))).... (-32.72 = -34.48 + 1.76)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:43:01 2006