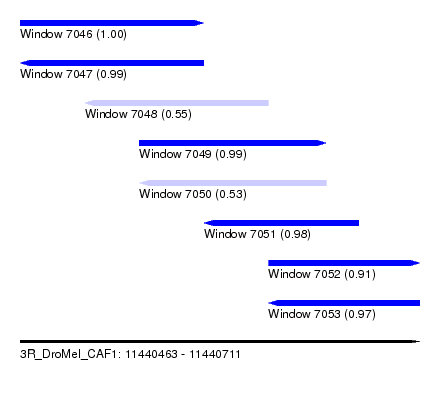
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 11,440,463 – 11,440,711 |
| Length | 248 |
| Max. P | 0.996077 |

| Location | 11,440,463 – 11,440,577 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.05 |
| Mean single sequence MFE | -37.50 |
| Consensus MFE | -32.72 |
| Energy contribution | -33.32 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.87 |
| SVM decision value | 2.65 |
| SVM RNA-class probability | 0.996077 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11440463 114 + 27905053 UGAGGGAAAGUCAAACAUGACGGACCCGACGAGUGGACAAGGACACUGACACCGG------CACAAGGACUCCCCCUAAAGCCGUGCAUCCAUGCACAGAAUGCAAUGGCAUAUUGCAAU ...(((...((((....))))...)))....((((........))))......((------(...(((......)))...)))(((((....)))))....(((((((...))))))).. ( -31.80) >DroSec_CAF1 180441 114 + 1 UGAGGGAAAGUCAAACAUGACGGACCCGACGAGUGGACAAGGACACUGACACCGG------CACAAGGACUCCCCCUAAAGCCGUGCAUCCAUGCACAGGAUGCAAUGGCAUAUUGCAAU ...(((...((((....))))...)))..((.(((....((....))..)))))(------((..(((......)))...(((((((((((.......)))))).)))))....)))... ( -36.80) >DroSim_CAF1 174631 114 + 1 UGAGGGAAAGUCAAACCUGACGGACCCGACGAGUGGACAAGGACACUGACACCGG------CACAAGGACUCCCCCUAAAGCCGUGCAUCCAUGCACAGGAUGCAAUGGCAUAUUGCAAU ...(((...((((....))))...)))..((.(((....((....))..)))))(------((..(((......)))...(((((((((((.......)))))).)))))....)))... ( -37.00) >DroEre_CAF1 169906 114 + 1 UGAGGGAAAGUCAAACAUGACGGAGCCGACGAGUGGACAAGGACACUGACACCGG------CACAAGGACUCCCCCCAAAGCCGUGCAUCCAUGCACAGGAUGCAAUGGCAUAUUGCAAU ...(((...((((....))))((((((..((.(((....((....))..))))).------.....)).)))).)))...(((((((((((.......)))))).))))).......... ( -39.50) >DroYak_CAF1 173314 120 + 1 UGAGGGAAAGUCAAACAUGACGGAGCCAACGAGUGGACAAGGACACUGACACCGGCAAUGGCACAAGGACUCCCCCUAAAGCCGUGCAUCCAUGCACAGGAUGCAAUGGCAUAUUGCAAU ..((((...((((....))))((((((..((.(((....((....))..)))))((....))....)).))))))))...(((((((((((.......)))))).))))).......... ( -42.40) >consensus UGAGGGAAAGUCAAACAUGACGGACCCGACGAGUGGACAAGGACACUGACACCGG______CACAAGGACUCCCCCUAAAGCCGUGCAUCCAUGCACAGGAUGCAAUGGCAUAUUGCAAU ..((((..((((........((....)).((.(((....((....))..))))).............))))..))))...(((((((((((.......)))))).))))).......... (-32.72 = -33.32 + 0.60)



| Location | 11,440,463 – 11,440,577 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.05 |
| Mean single sequence MFE | -39.78 |
| Consensus MFE | -37.82 |
| Energy contribution | -37.54 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.95 |
| SVM decision value | 2.04 |
| SVM RNA-class probability | 0.986260 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11440463 114 - 27905053 AUUGCAAUAUGCCAUUGCAUUCUGUGCAUGGAUGCACGGCUUUAGGGGGAGUCCUUGUG------CCGGUGUCAGUGUCCUUGUCCACUCGUCGGGUCCGUCAUGUUUGACUUUCCCUCA ..((((((.....))))))..(((((((....)))))))....((((..((((..((((------.(((((.(((((........)))).).))...))).))))...))))..)))).. ( -36.50) >DroSec_CAF1 180441 114 - 1 AUUGCAAUAUGCCAUUGCAUCCUGUGCAUGGAUGCACGGCUUUAGGGGGAGUCCUUGUG------CCGGUGUCAGUGUCCUUGUCCACUCGUCGGGUCCGUCAUGUUUGACUUUCCCUCA ..........(((..(((((((.......))))))).)))...((((..((((..((((------.(((((.(((((........)))).).))...))).))))...))))..)))).. ( -38.80) >DroSim_CAF1 174631 114 - 1 AUUGCAAUAUGCCAUUGCAUCCUGUGCAUGGAUGCACGGCUUUAGGGGGAGUCCUUGUG------CCGGUGUCAGUGUCCUUGUCCACUCGUCGGGUCCGUCAGGUUUGACUUUCCCUCA ..........(((..(((((((.......))))))).)))...((((..((((((((((------(((..(..((((........)))))..)))...)).))))...))))..)))).. ( -39.20) >DroEre_CAF1 169906 114 - 1 AUUGCAAUAUGCCAUUGCAUCCUGUGCAUGGAUGCACGGCUUUGGGGGGAGUCCUUGUG------CCGGUGUCAGUGUCCUUGUCCACUCGUCGGCUCCGUCAUGUUUGACUUUCCCUCA ....(((...(((..(((((((.......))))))).))).)))(((..((((..((((------.(((.(((((((........))))....))).))).))))...))))..)))... ( -42.90) >DroYak_CAF1 173314 120 - 1 AUUGCAAUAUGCCAUUGCAUCCUGUGCAUGGAUGCACGGCUUUAGGGGGAGUCCUUGUGCCAUUGCCGGUGUCAGUGUCCUUGUCCACUCGUUGGCUCCGUCAUGUUUGACUUUCCCUCA ..........(((..(((((((.......))))))).)))...((((..((((..((((.(...(((..((..((((........))))))..)))...).))))...))))..)))).. ( -41.50) >consensus AUUGCAAUAUGCCAUUGCAUCCUGUGCAUGGAUGCACGGCUUUAGGGGGAGUCCUUGUG______CCGGUGUCAGUGUCCUUGUCCACUCGUCGGGUCCGUCAUGUUUGACUUUCCCUCA ..........(((..(((((((.......))))))).)))...((((..((((..((((......(((..(..((((........)))))..)))......))))...))))..)))).. (-37.82 = -37.54 + -0.28)



| Location | 11,440,503 – 11,440,617 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.22 |
| Mean single sequence MFE | -38.87 |
| Consensus MFE | -35.16 |
| Energy contribution | -35.04 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.17 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.546562 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11440503 114 - 27905053 ACAUGCCGCAAGUCUCGCCAACCACUUGGACCUCCAACUAAUUGCAAUAUGCCAUUGCAUUCUGUGCAUGGAUGCACGGCUUUAGGGGGAGUCCUUGUG------CCGGUGUCAGUGUCC .......(((.....((((...(((..((((.(((..((((.((((((.....))))))..(((((((....)))))))..))))..)))))))..)))------..))))....))).. ( -38.90) >DroSec_CAF1 180481 114 - 1 ACAUGCCGCAAGUCUCGCCAACCACUUGGAGCUCCAACUAAUUGCAAUAUGCCAUUGCAUCCUGUGCAUGGAUGCACGGCUUUAGGGGGAGUCCUUGUG------CCGGUGUCAGUGUCC .......(((.....((((...(((..(((.((((..((((.........(((..(((((((.......))))))).))).))))..)))))))..)))------..))))....))).. ( -39.50) >DroSim_CAF1 174671 114 - 1 ACAUGCCGCAAGUCUCGCCAACCACUUGGACCUCCAACUAAUUGCAAUAUGCCAUUGCAUCCUGUGCAUGGAUGCACGGCUUUAGGGGGAGUCCUUGUG------CCGGUGUCAGUGUCC .......(((.....((((...(((..((((.(((..((((.........(((..(((((((.......))))))).))).))))..)))))))..)))------..))))....))).. ( -39.50) >DroEre_CAF1 169946 114 - 1 ACAUGCCGCAAGUCCCGCCAACCACUUGGACCGCCAACUAAUUGCAAUAUGCCAUUGCAUCCUGUGCAUGGAUGCACGGCUUUGGGGGGAGUCCUUGUG------CCGGUGUCAGUGUCC (((((.((((((((((.((((((....))...(((........((.....))...(((((((.......))))))).))).)))).))))...))))))------.).))))........ ( -38.20) >DroYak_CAF1 173354 120 - 1 ACAUGCCGCAAGUCUCGCCAACCACUUGGACCUCCAACUAAUUGCAAUAUGCCAUUGCAUCCUGUGCAUGGAUGCACGGCUUUAGGGGGAGUCCUUGUGCCAUUGCCGGUGUCAGUGUCC ..(((((((((...........(((..((((.(((..((((.........(((..(((((((.......))))))).))).))))..)))))))..)))...))).))))))........ ( -38.24) >consensus ACAUGCCGCAAGUCUCGCCAACCACUUGGACCUCCAACUAAUUGCAAUAUGCCAUUGCAUCCUGUGCAUGGAUGCACGGCUUUAGGGGGAGUCCUUGUG______CCGGUGUCAGUGUCC ..(((((((.......))....(((..(((.((((..((((.........(((..(((((((.......))))))).))).))))..)))))))..)))........)))))........ (-35.16 = -35.04 + -0.12)


| Location | 11,440,537 – 11,440,653 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.54 |
| Mean single sequence MFE | -44.72 |
| Consensus MFE | -41.18 |
| Energy contribution | -41.06 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.92 |
| SVM decision value | 2.15 |
| SVM RNA-class probability | 0.989083 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11440537 116 + 27905053 GCCGUGCAUCCAUGCACAGAAUGCAAUGGCAUAUUGCAAUUAGUUGGAGGUCCAAGUGGUUGGCGAGACUUGCGGCAUGUGUGGAGGGCUAAUAAGCUGGGUGGCGGGGAUUCCCG---- .((((.(((((.(.((((..((((....(((..(((((((((.((((....)))).))))).))))....))).)))).)))).).((((....)))))))))))))((....)).---- ( -41.20) >DroSec_CAF1 180515 116 + 1 GCCGUGCAUCCAUGCACAGGAUGCAAUGGCAUAUUGCAAUUAGUUGGAGCUCCAAGUGGUUGGCGAGACUUGCGGCAUGUGUGGAGGGCUAAUAAGCUGGGUGGCGGGGAUUCCCU---- (((.(.((..(((((.((((((((....)))).(((((((((.((((....)))).))))).))))..))))..)))))..)).).)))......(((....)))(((....))).---- ( -42.60) >DroSim_CAF1 174705 116 + 1 GCCGUGCAUCCAUGCACAGGAUGCAAUGGCAUAUUGCAAUUAGUUGGAGGUCCAAGUGGUUGGCGAGACUUGCGGCAUGUGUGGAGGGCUAAUAAGCUGGGUGGCGGGGACUCCCG---- (((.(.((..(((((.((((((((....)))).(((((((((.((((....)))).))))).))))..))))..)))))..)).).)))......(((....)))(((....))).---- ( -43.10) >DroEre_CAF1 169980 120 + 1 GCCGUGCAUCCAUGCACAGGAUGCAAUGGCAUAUUGCAAUUAGUUGGCGGUCCAAGUGGUUGGCGGGACUUGCGGCAUGUGUGGAGGGCUAAUAAGCUGGGUGGCGGGGAUUCUCGCCAG (((((((((((.......)))))).))))).......(((((.((((....)))).)))))((((((((((((.((..((.(((....)))....))...)).)))))...))))))).. ( -45.60) >DroYak_CAF1 173394 120 + 1 GCCGUGCAUCCAUGCACAGGAUGCAAUGGCAUAUUGCAAUUAGUUGGAGGUCCAAGUGGUUGGCGAGACUUGCGGCAUGUGUGGAGGGCUAAUAAGCUGGGUGGCGGGGAUUCCCGCCAU (((.(.((..(((((.((((((((....)))).(((((((((.((((....)))).))))).))))..))))..)))))..)).).)))...........((((((((....)))))))) ( -51.10) >consensus GCCGUGCAUCCAUGCACAGGAUGCAAUGGCAUAUUGCAAUUAGUUGGAGGUCCAAGUGGUUGGCGAGACUUGCGGCAUGUGUGGAGGGCUAAUAAGCUGGGUGGCGGGGAUUCCCG____ (((.(.((..(((((.((((((((....)))).(((((((((.((((....)))).))))).))))..))))..)))))..)).).)))......(((....)))(((....)))..... (-41.18 = -41.06 + -0.12)



| Location | 11,440,537 – 11,440,653 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.54 |
| Mean single sequence MFE | -30.22 |
| Consensus MFE | -26.00 |
| Energy contribution | -26.20 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.86 |
| SVM decision value | -0.00 |
| SVM RNA-class probability | 0.531900 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11440537 116 - 27905053 ----CGGGAAUCCCCGCCACCCAGCUUAUUAGCCCUCCACACAUGCCGCAAGUCUCGCCAACCACUUGGACCUCCAACUAAUUGCAAUAUGCCAUUGCAUUCUGUGCAUGGAUGCACGGC ----.(((....)))(((.....(((....)))....((..(((((.(((...............((((....)))).....((((((.....))))))...))))))))..))...))) ( -27.70) >DroSec_CAF1 180515 116 - 1 ----AGGGAAUCCCCGCCACCCAGCUUAUUAGCCCUCCACACAUGCCGCAAGUCUCGCCAACCACUUGGAGCUCCAACUAAUUGCAAUAUGCCAUUGCAUCCUGUGCAUGGAUGCACGGC ----.(((....)))(((.....((..(((((..(((((........((.......))........)))))......))))).))..........(((((((.......))))))).))) ( -29.09) >DroSim_CAF1 174705 116 - 1 ----CGGGAGUCCCCGCCACCCAGCUUAUUAGCCCUCCACACAUGCCGCAAGUCUCGCCAACCACUUGGACCUCCAACUAAUUGCAAUAUGCCAUUGCAUCCUGUGCAUGGAUGCACGGC ----.(((....)))(((.....((..(((((..............(....).............((((....))))))))).))..........(((((((.......))))))).))) ( -28.00) >DroEre_CAF1 169980 120 - 1 CUGGCGAGAAUCCCCGCCACCCAGCUUAUUAGCCCUCCACACAUGCCGCAAGUCCCGCCAACCACUUGGACCGCCAACUAAUUGCAAUAUGCCAUUGCAUCCUGUGCAUGGAUGCACGGC .(((((........)))))....(((....)))...........(((((((.....((...((....))...)).......))))..........(((((((.......))))))).))) ( -29.70) >DroYak_CAF1 173394 120 - 1 AUGGCGGGAAUCCCCGCCACCCAGCUUAUUAGCCCUCCACACAUGCCGCAAGUCUCGCCAACCACUUGGACCUCCAACUAAUUGCAAUAUGCCAUUGCAUCCUGUGCAUGGAUGCACGGC .(((((((....)))))))....((..(((((..............(....).............((((....))))))))).)).....(((..(((((((.......))))))).))) ( -36.60) >consensus ____CGGGAAUCCCCGCCACCCAGCUUAUUAGCCCUCCACACAUGCCGCAAGUCUCGCCAACCACUUGGACCUCCAACUAAUUGCAAUAUGCCAUUGCAUCCUGUGCAUGGAUGCACGGC .....(((....)))(((.....(((....)))....((..(((((.(((...............((((....)))).....((((((.....))))))...))))))))..))...))) (-26.00 = -26.20 + 0.20)



| Location | 11,440,577 – 11,440,673 |
|---|---|
| Length | 96 |
| Sequences | 4 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 94.00 |
| Mean single sequence MFE | -36.60 |
| Consensus MFE | -32.22 |
| Energy contribution | -32.48 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.75 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.85 |
| SVM RNA-class probability | 0.979959 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11440577 96 - 27905053 AUAACAGGCCGGAGGGCUGA--------CGGGAAUCCCCGCCACCCAGCUUAUUAGCCCUCCACACAUGCCGCAAGUCUCGCCAACCACUUGGACCUCCAACUA ......(((.((((((((((--------.(((....)))((......))...))))))))))........(....)....)))......((((....))))... ( -33.40) >DroSec_CAF1 180555 96 - 1 AUAACAGGCCGGAGGGCUGA--------AGGGAAUCCCCGCCACCCAGCUUAUUAGCCCUCCACACAUGCCGCAAGUCUCGCCAACCACUUGGAGCUCCAACUA ......(((.((((((((((--------.(((....)))((......))...))))))))))........(....)....)))......((((....))))... ( -33.60) >DroSim_CAF1 174745 96 - 1 AUAACAGGCCGGAGGGCUGA--------CGGGAGUCCCCGCCACCCAGCUUAUUAGCCCUCCACACAUGCCGCAAGUCUCGCCAACCACUUGGACCUCCAACUA ......(((.((((((((((--------.(((....)))((......))...))))))))))........(....)....)))......((((....))))... ( -33.40) >DroYak_CAF1 173434 104 - 1 AUAACAGGCCGGAGGGCUGUCGGGAUGGCGGGAAUCCCCGCCACCCAGCUUAUUAGCCCUCCACACAUGCCGCAAGUCUCGCCAACCACUUGGACCUCCAACUA ......(((.((((((((((.(((.(((((((....)))))))))).)).....))))))))........(....)....)))......((((....))))... ( -46.00) >consensus AUAACAGGCCGGAGGGCUGA________CGGGAAUCCCCGCCACCCAGCUUAUUAGCCCUCCACACAUGCCGCAAGUCUCGCCAACCACUUGGACCUCCAACUA ......(((.((((((((((.........(((....)))((......))...))))))))))........(....)....)))......((((....))))... (-32.22 = -32.48 + 0.25)



| Location | 11,440,617 – 11,440,711 |
|---|---|
| Length | 94 |
| Sequences | 3 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 81.94 |
| Mean single sequence MFE | -34.97 |
| Consensus MFE | -28.10 |
| Energy contribution | -28.10 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.10 |
| SVM RNA-class probability | 0.914754 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11440617 94 + 27905053 GUGGAGGGCUAAUAAGCUGGGUGGCGGGGAUUCCCG--------UCAGCCCUCCGGCCUGUUAUCCUUUUGCCUGAACACACACACACACACGAACAUACUC (..(((((.(((((.(((((((((((((....))))--------)))....)))))).))))).)))))..).............................. ( -32.20) >DroSim_CAF1 174785 84 + 1 GUGGAGGGCUAAUAAGCUGGGUGGCGGGGACUCCCG--------UCAGCCCUCCGGCCUGUUAUCCUUUGGCCAGGACACACACA----------CACACUC ..((((((((....))))((.(((((((....))))--------))).))))))((((...........))))............----------....... ( -32.80) >DroYak_CAF1 173474 90 + 1 GUGGAGGGCUAAUAAGCUGGGUGGCGGGGAUUCCCGCCAUCCCGACAGCCCUCCGGCCUGUUAUCCUUUUGCCUGGACACACA------------CACACUC .(((((((((......(.((((((((((....)))))))))).)..)))))))))...(((..(((........)))...)))------------....... ( -39.90) >consensus GUGGAGGGCUAAUAAGCUGGGUGGCGGGGAUUCCCG________UCAGCCCUCCGGCCUGUUAUCCUUUUGCCUGGACACACACA__________CACACUC (..(((((.(((((.((((((.((((((....)))(.........).))).)))))).))))).)))))..).............................. (-28.10 = -28.10 + -0.00)



| Location | 11,440,617 – 11,440,711 |
|---|---|
| Length | 94 |
| Sequences | 3 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 81.94 |
| Mean single sequence MFE | -39.20 |
| Consensus MFE | -29.65 |
| Energy contribution | -29.77 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.62 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.73 |
| SVM RNA-class probability | 0.974480 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11440617 94 - 27905053 GAGUAUGUUCGUGUGUGUGUGUGUGUUCAGGCAAAAGGAUAACAGGCCGGAGGGCUGA--------CGGGAAUCCCCGCCACCCAGCUUAUUAGCCCUCCAC ..((.(((((...(((.((........)).)))...))))))).....((((((((((--------.(((....)))((......))...)))))))))).. ( -33.80) >DroSim_CAF1 174785 84 - 1 GAGUGUG----------UGUGUGUGUCCUGGCCAAAGGAUAACAGGCCGGAGGGCUGA--------CGGGAGUCCCCGCCACCCAGCUUAUUAGCCCUCCAC ....(.(----------(.(((.((((((......))))))))).)))((((((((((--------.(((....)))((......))...)))))))))).. ( -36.60) >DroYak_CAF1 173474 90 - 1 GAGUGUG------------UGUGUGUCCAGGCAAAAGGAUAACAGGCCGGAGGGCUGUCGGGAUGGCGGGAAUCCCCGCCACCCAGCUUAUUAGCCCUCCAC ..(.((.------------(((.(((((........)))))))).)))((((((((((.(((.(((((((....)))))))))).)).....)))))))).. ( -47.20) >consensus GAGUGUG__________UGUGUGUGUCCAGGCAAAAGGAUAACAGGCCGGAGGGCUGA________CGGGAAUCCCCGCCACCCAGCUUAUUAGCCCUCCAC ..................((.(((((((........)))).))).)).(((((((((..........(((....)))((......))....))))))))).. (-29.65 = -29.77 + 0.11)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:26:00 2006