
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
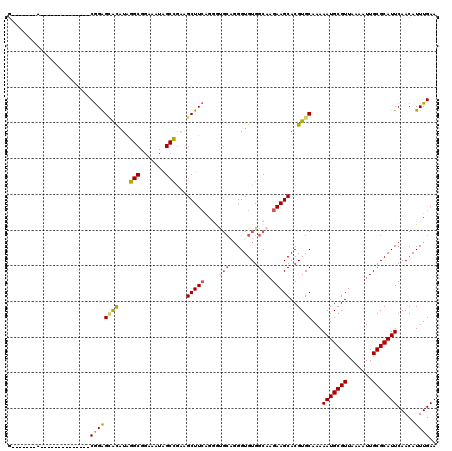
| Location | 11,437,127 – 11,437,306 |
| Length | 179 |
| Max. P | 0.997228 |

| Location | 11,437,127 – 11,437,226 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.62 |
| Mean single sequence MFE | -28.40 |
| Consensus MFE | -22.28 |
| Energy contribution | -22.40 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.578663 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11437127 99 + 27905053 G-------A--------------CGGAGCACAUAGGCGGAAAUAGCCGGAGCUUCGGGGUGCAGGGUGUGGCAAGAAGCACGUGCAAAAAUGCGUUAAAAUUGCGCAUUCAACAUUUGAA .-------.--------------((((((((...(((.......)))...(((((..(.(((.....))).)..)))))..))))...(((((((.......))))))).....)))).. ( -30.70) >DroSec_CAF1 177213 92 + 1 G-------A--------------CGGAGCGCAUAGGCGGAAAUAGCCGAAGCUUCAG-------GGUGUGGCACGAAGCACGUGCAAAAAUGCGUUAAAAUUGCGCAUUCAACAUUUGAA .-------.--------------.(((((.....(((.......)))...))))).(-------(((((.(((((.....)))))...(((((((.......)))))))..))))))... ( -29.80) >DroSim_CAF1 171410 99 + 1 G-------G--------------CGGAGCACAUAGGCGGAAAUAGCCGAAGCUUCAGCGUGCUGGGUGUGGCAAGAAGCACGUGCAAAAAUGCGUUAAAAUUGCGCAUUCAACAUUUGAA .-------(--------------((((((.....(((.......)))...))))).(((((((...((...))...)))))))))...(((((((.......)))))))........... ( -30.90) >DroEre_CAF1 166786 100 + 1 U-------A-------------CCGGGGGAUAUGGGCGGAAAUAGCUUAGGCUUCAGGACGCAGGGUGUGGCAAGAAGCACGUGCAAAAAUGCGUUAAAAUUGCGCAUUCAACAUUUGAA .-------.-------------((.((((...(((((.......)))))..)))).))..(((.(.(((........)))).)))...(((((((.......)))))))........... ( -25.00) >DroYak_CAF1 169662 120 + 1 GGGAGGGGAGCGGAGGUGAGGGGCAGGGUAUAUAGGCGGAAAUGGCCGAAGCUUGAGGGUGCAGGGUGUGGCAAGAAGCACGUGCAAAAAUGCGUUAAAAUUGCGCAUUCAACAUUUGAA ..........((((..((((..(((.(.......(((.......)))...((((...(.(((.....))).)...)))).).))).....(((((.......)))))))))...)))).. ( -25.60) >consensus G_______A______________CGGAGCACAUAGGCGGAAAUAGCCGAAGCUUCAGGGUGCAGGGUGUGGCAAGAAGCACGUGCAAAAAUGCGUUAAAAUUGCGCAUUCAACAUUUGAA .......................((((((((...(((.......)))...(((((....(((........))).)))))..))))...(((((((.......))))))).....)))).. (-22.28 = -22.40 + 0.12)


| Location | 11,437,146 – 11,437,266 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.67 |
| Mean single sequence MFE | -34.12 |
| Consensus MFE | -29.36 |
| Energy contribution | -30.04 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.73 |
| SVM RNA-class probability | 0.974213 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11437146 120 + 27905053 AAUAGCCGGAGCUUCGGGGUGCAGGGUGUGGCAAGAAGCACGUGCAAAAAUGCGUUAAAAUUGCGCAUUCAACAUUUGAACGCAAACACAAACGCAAUUAAGCGCGGCGUCAGUGUAAUG ....(((...(((((..(.(((.....))).)..)))))..((((..((.((((((....(((((..((((.....))))))))).....)))))).))..)))))))............ ( -35.60) >DroSec_CAF1 177232 113 + 1 AAUAGCCGAAGCUUCAG-------GGUGUGGCACGAAGCACGUGCAAAAAUGCGUUAAAAUUGCGCAUUCAACAUUUGAACGCAAACACAAACGCAAUUAAGCGCGGCGUCAGUGUAAUG ....((((..(((((((-------(.(((.(((((.....)))))...(((((((.......)))))))..))))))))).)).........(((......)))))))............ ( -32.90) >DroSim_CAF1 171429 120 + 1 AAUAGCCGAAGCUUCAGCGUGCUGGGUGUGGCAAGAAGCACGUGCAAAAAUGCGUUAAAAUUGCGCAUUCAACAUUUGAACCCAAACACAAACGCAAUUAAGCGCGGCGUCAGUGUAAUG ....((((..((((..((((..(((((....((((..((....))...(((((((.......))))))).....)))).))))).......))))....)))).))))............ ( -32.20) >DroEre_CAF1 166806 120 + 1 AAUAGCUUAGGCUUCAGGACGCAGGGUGUGGCAAGAAGCACGUGCAAAAAUGCGUUAAAAUUGCGCAUUCAACAUUUGAACGCAAACACAAACGCAAUUAAGCGCGGCGUCAGUGUAAUG ......(((.(((....(((((...((((........))))((((..((.((((((....(((((..((((.....))))))))).....)))))).))..)))).)))))))).))).. ( -33.30) >DroYak_CAF1 169702 120 + 1 AAUGGCCGAAGCUUGAGGGUGCAGGGUGUGGCAAGAAGCACGUGCAAAAAUGCGUUAAAAUUGCGCAUUCAACAUUUGAACGCAAACACAAACGCAAUUAAGCGCGGCGUCAGUGUAAUG ....((((..((((((...(((.(..((((((((.....((((........)))).....))))((.((((.....)))).))...))))..)))).)))))).))))............ ( -36.60) >consensus AAUAGCCGAAGCUUCAGGGUGCAGGGUGUGGCAAGAAGCACGUGCAAAAAUGCGUUAAAAUUGCGCAUUCAACAUUUGAACGCAAACACAAACGCAAUUAAGCGCGGCGUCAGUGUAAUG ....((((.(.(((........))).).))))..((.((.(((((..((.((((((....(((((..((((.....))))))))).....)))))).))..))))))).))......... (-29.36 = -30.04 + 0.68)


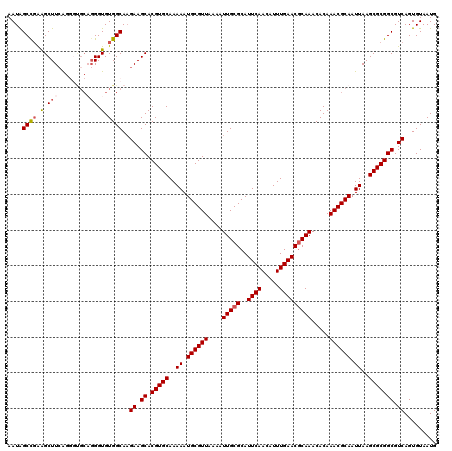
| Location | 11,437,146 – 11,437,266 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.67 |
| Mean single sequence MFE | -33.66 |
| Consensus MFE | -29.46 |
| Energy contribution | -29.86 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.88 |
| SVM decision value | 2.82 |
| SVM RNA-class probability | 0.997228 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11437146 120 - 27905053 CAUUACACUGACGCCGCGCUUAAUUGCGUUUGUGUUUGCGUUCAAAUGUUGAAUGCGCAAUUUUAACGCAUUUUUGCACGUGCUUCUUGCCACACCCUGCACCCCGAAGCUCCGGCUAUU ............((((.((..((.((((((...(((((((((((.....)))))))).)))...)))))).))..))..(.(((((.(((........)))....))))).))))).... ( -34.90) >DroSec_CAF1 177232 113 - 1 CAUUACACUGACGCCGCGCUUAAUUGCGUUUGUGUUUGCGUUCAAAUGUUGAAUGCGCAAUUUUAACGCAUUUUUGCACGUGCUUCGUGCCACACC-------CUGAAGCUUCGGCUAUU .....(((.((.((((.((..((.((((((...(((((((((((.....)))))))).)))...)))))).))..)).)).)).))))).......-------....(((....)))... ( -35.30) >DroSim_CAF1 171429 120 - 1 CAUUACACUGACGCCGCGCUUAAUUGCGUUUGUGUUUGGGUUCAAAUGUUGAAUGCGCAAUUUUAACGCAUUUUUGCACGUGCUUCUUGCCACACCCAGCACGCUGAAGCUUCGGCUAUU ............((.((((......))))..(((((.((((.....(((.(((((((.........)))))))..))).((((.....).))))))))))))))...(((....)))... ( -33.30) >DroEre_CAF1 166806 120 - 1 CAUUACACUGACGCCGCGCUUAAUUGCGUUUGUGUUUGCGUUCAAAUGUUGAAUGCGCAAUUUUAACGCAUUUUUGCACGUGCUUCUUGCCACACCCUGCGUCCUGAAGCCUAAGCUAUU .........(((((.(.((..((.((((((...(((((((((((.....)))))))).)))...)))))).))..)).)((((.....).))).....)))))....(((....)))... ( -32.80) >DroYak_CAF1 169702 120 - 1 CAUUACACUGACGCCGCGCUUAAUUGCGUUUGUGUUUGCGUUCAAAUGUUGAAUGCGCAAUUUUAACGCAUUUUUGCACGUGCUUCUUGCCACACCCUGCACCCUCAAGCUUCGGCCAUU .........((.((((.((..((.((((((...(((((((((((.....)))))))).)))...)))))).))..)).)).)).)).(((........))).......((....)).... ( -32.00) >consensus CAUUACACUGACGCCGCGCUUAAUUGCGUUUGUGUUUGCGUUCAAAUGUUGAAUGCGCAAUUUUAACGCAUUUUUGCACGUGCUUCUUGCCACACCCUGCACCCUGAAGCUUCGGCUAUU .........((.((((.((..((.((((((...(((((((((((.....)))))))).)))...)))))).))..)).)).)).)).....................(((....)))... (-29.46 = -29.86 + 0.40)



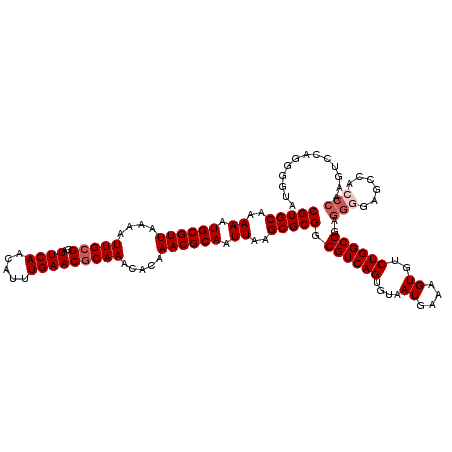
| Location | 11,437,186 – 11,437,306 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.07 |
| Mean single sequence MFE | -35.10 |
| Consensus MFE | -27.18 |
| Energy contribution | -30.18 |
| Covariance contribution | 3.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.87 |
| SVM RNA-class probability | 0.870154 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11437186 120 + 27905053 CGUGCAAAAAUGCGUUAAAAUUGCGCAUUCAACAUUUGAACGCAAACACAAACGCAAUUAAGCGCGGCGUCAGUGUAAUGAAAGUGUCUGGCGGAGGGGAGCCACCCAAGUCCAAGGGUA (((((..((.((((((....(((((..((((.....))))))))).....)))))).))..))))).((((((....((....))..))))))..((....))((((........)))). ( -37.00) >DroSec_CAF1 177265 120 + 1 CGUGCAAAAAUGCGUUAAAAUUGCGCAUUCAACAUUUGAACGCAAACACAAACGCAAUUAAGCGCGGCGUCAGUGUAAUGAAAGUGUCUGGCGGAGGGGAGCCACCCAAGUCCAGGGGUA (((((..((.((((((....(((((..((((.....))))))))).....)))))).))..))))).((((((....((....))..))))))..((....))((((........)))). ( -36.90) >DroSim_CAF1 171469 120 + 1 CGUGCAAAAAUGCGUUAAAAUUGCGCAUUCAACAUUUGAACCCAAACACAAACGCAAUUAAGCGCGGCGUCAGUGUAAUGAAAGUGUCUGGCGGAGGGGAGCCACCCAAGUCCAGGGGUA .((.....(((((((.......)))))))..))......((((..((((..((((...........))))..))))...........(((((...(((......)))..).)))))))). ( -33.00) >DroEre_CAF1 166846 118 + 1 CGUGCAAAAAUGCGUUAAAAUUGCGCAUUCAACAUUUGAACGCAAACACAAACGCAAUUAAGCGCGGCGUCAGUGUAAUGAAAGUGUCUGGCGGGGGC--GCCACCCAAGGCCGGGCGAA (((((..((.((((((....(((((..((((.....))))))))).....)))))).))..)))))...(((......)))...((((((((.(((..--....)))...)))))))).. ( -39.30) >DroYak_CAF1 169742 98 + 1 CGUGCAAAAAUGCGUUAAAAUUGCGCAUUCAACAUUUGAACGCAAACACAAACGCAAUUAAGCGCGGCGUCAGUGUAAUGAAAGUGUCUGGCGGAGGG---------------------- (((((..((.((((((....(((((..((((.....))))))))).....)))))).))..))))).((((((....((....))..)))))).....---------------------- ( -29.30) >consensus CGUGCAAAAAUGCGUUAAAAUUGCGCAUUCAACAUUUGAACGCAAACACAAACGCAAUUAAGCGCGGCGUCAGUGUAAUGAAAGUGUCUGGCGGAGGGGAGCCACCCAAGUCCAGGGGUA (((((..((.((((((....(((((..((((.....))))))))).....)))))).))..))))).((((((....((....))..))))))..(((......)))............. (-27.18 = -30.18 + 3.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:25:47 2006