
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 11,434,800 – 11,435,017 |
| Length | 217 |
| Max. P | 0.999776 |

| Location | 11,434,800 – 11,434,915 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.10 |
| Mean single sequence MFE | -31.30 |
| Consensus MFE | -27.84 |
| Energy contribution | -27.88 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.89 |
| SVM decision value | 2.15 |
| SVM RNA-class probability | 0.989057 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11434800 115 - 27905053 CUAUCAGCGAAAUGAAAUUGAUUGGAAAAUGCCAAUGAAAAGAUUGCAUUGUGCAGCUAUUAAGCGGGGGAAUAGUUCUUCGCUAAGUGGUCGGGCGAAUGCACUUU-----UUCAGAAG ...(((......))).....(((((......)))))((((((..((((((((.(.((((((.(((((((((....))))))))).)))))).).)).)))))).)))-----)))..... ( -34.10) >DroSec_CAF1 174899 120 - 1 CUAUCAGCGAAAUGAAAUUGAUUGGAAAAUGCCAAUGAAAAGAUUGCAUUGUGCAGCUAUUAAGCGGGGGAAUAGUUCUUCGCUAAGUGGUCGGGCGAAUGCACUUAUGGUAUUCAGAAG .(((((...........((.(((((......))))).)).....((((((((.(.((((((.(((((((((....))))))))).)))))).).)).))))))....)))))........ ( -33.60) >DroSim_CAF1 169115 115 - 1 CUAUCAGCGAAAUGAAAUUGAUUGGAAAAUGCCAAUGAAAAGAUUGCAUUGUUCAGCUAUUAAGCGGGGGAAUAGUUCUUCGCUAAGUGGUCGGGCGAAUGCACUUA-----UUCAGAAG ............((((.((.(((((......))))).)).....((((((((((.((((((.(((((((((....))))))))).)))))).)))).))))))....-----)))).... ( -34.10) >DroEre_CAF1 164477 115 - 1 CUAUCAGCGAAAUGAAAUUGAUUGGAAAAUGCCAAUGAAAAGAUUGCAUUGUGUAGCUAUUAAGCGGGGGAAUAGUUCCACGCUAAGUGCUCGGUUGAAUGCACUUU-----UUCAGAAG ...(((......))).....(((((......)))))((((((..((((((.((.(((.(((.((((.((((....)))).)))).))))))))....)))))).)))-----)))..... ( -25.70) >DroYak_CAF1 167229 115 - 1 CUAUCAGCGAAAUGCAAUUGAUUGGAAAAUGCCAAUGAAAAGAUUGCAUUGUGUAGCUAUUAAGCGGGCGAAUAGUUCUUCGCUAAGUGGUCGGAUGAAUGCACUUA-----UUCAGAAG ......(((.(((((((((.(((((......))))).....))))))))).))).((((((.((((((.((.....)))))))).)))))).((((((......)))-----)))..... ( -29.00) >consensus CUAUCAGCGAAAUGAAAUUGAUUGGAAAAUGCCAAUGAAAAGAUUGCAUUGUGCAGCUAUUAAGCGGGGGAAUAGUUCUUCGCUAAGUGGUCGGGCGAAUGCACUUA_____UUCAGAAG ........(((......((.(((((......))))).)).....((((((((.(.((((((.(((((((((....))))))))).)))))).).)).)))))).........)))..... (-27.84 = -27.88 + 0.04)



| Location | 11,434,835 – 11,434,955 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.17 |
| Mean single sequence MFE | -30.18 |
| Consensus MFE | -24.80 |
| Energy contribution | -25.84 |
| Covariance contribution | 1.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.785880 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11434835 120 + 27905053 AAGAACUAUUCCCCCGCUUAAUAGCUGCACAAUGCAAUCUUUUCAUUGGCAUUUUCCAAUCAAUUUCAUUUCGCUGAUAGUGGCAGUUAGCGAAGCUGGCGAAGCCAUCAAACUGGCAAA ........(((.((.((((..(((((((.(((((.((....))))))).......................(((.....))))))))))...)))).)).)))((((......))))... ( -30.30) >DroSec_CAF1 174939 120 + 1 AAGAACUAUUCCCCCGCUUAAUAGCUGCACAAUGCAAUCUUUUCAUUGGCAUUUUCCAAUCAAUUUCAUUUCGCUGAUAGUGGCAGUUAGCGGAGCUGGCGAAGCCAUCAAACUGGCAAA ........(((.((.((((..(((((((.(((((.((....))))))).......................(((.....))))))))))...)))).)).)))((((......))))... ( -30.70) >DroSim_CAF1 169150 120 + 1 AAGAACUAUUCCCCCGCUUAAUAGCUGAACAAUGCAAUCUUUUCAUUGGCAUUUUCCAAUCAAUUUCAUUUCGCUGAUAGUGGCAGUUAGCGUAGCUGGAGAAGCCAUCAAACUGGCAAA ........(((.((.(((.....(((((....(((.........(((((......)))))...........(((.....)))))).)))))..))).)).)))((((......))))... ( -26.70) >DroEre_CAF1 164512 120 + 1 UGGAACUAUUCCCCCGCUUAAUAGCUACACAAUGCAAUCUUUUCAUUGGCAUUUUCCAAUCAAUUUCAUUUCGCUGAUAGUGGCAGUUAGCGGAGCUGGCGAAGCCAUCAAACUGGCAAA ........((((((((((.(((.(((((.(((((.((....)))))))..........((((............)))).))))).))))))))....)).)))((((......))))... ( -27.50) >DroYak_CAF1 167264 120 + 1 AAGAACUAUUCGCCCGCUUAAUAGCUACACAAUGCAAUCUUUUCAUUGGCAUUUUCCAAUCAAUUGCAUUUCGCUGAUAGUGGCAGUUAGCGAAGCUGGCGUUGCCAUCAAACUGGCAAA ..........((((.((((....((((.((((((((((......(((((......)))))..))))))))..(((......))).)))))).)))).))))((((((......)))))). ( -35.70) >consensus AAGAACUAUUCCCCCGCUUAAUAGCUGCACAAUGCAAUCUUUUCAUUGGCAUUUUCCAAUCAAUUUCAUUUCGCUGAUAGUGGCAGUUAGCGAAGCUGGCGAAGCCAUCAAACUGGCAAA ........(((.((.((((..(((((((.(((((.((....))))))).......................(((.....))))))))))...)))).)).)))((((......))))... (-24.80 = -25.84 + 1.04)



| Location | 11,434,835 – 11,434,955 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.17 |
| Mean single sequence MFE | -32.78 |
| Consensus MFE | -28.34 |
| Energy contribution | -28.94 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.570567 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11434835 120 - 27905053 UUUGCCAGUUUGAUGGCUUCGCCAGCUUCGCUAACUGCCACUAUCAGCGAAAUGAAAUUGAUUGGAAAAUGCCAAUGAAAAGAUUGCAUUGUGCAGCUAUUAAGCGGGGGAAUAGUUCUU (((.((.(((((((((((.(((..((((((((.............))))))......((.(((((......))))).))......))...))).))))))))))).)).)))........ ( -33.02) >DroSec_CAF1 174939 120 - 1 UUUGCCAGUUUGAUGGCUUCGCCAGCUCCGCUAACUGCCACUAUCAGCGAAAUGAAAUUGAUUGGAAAAUGCCAAUGAAAAGAUUGCAUUGUGCAGCUAUUAAGCGGGGGAAUAGUUCUU (((.((.(((((((((((.(((..((..((((.............))))........((.(((((......))))).))......))...))).))))))))))).)).)))........ ( -30.72) >DroSim_CAF1 169150 120 - 1 UUUGCCAGUUUGAUGGCUUCUCCAGCUACGCUAACUGCCACUAUCAGCGAAAUGAAAUUGAUUGGAAAAUGCCAAUGAAAAGAUUGCAUUGUUCAGCUAUUAAGCGGGGGAAUAGUUCUU ...((((......))))((((((.(((.((((.............))))............(((((.(((((.(((......)))))))).)))))......))).))))))........ ( -27.32) >DroEre_CAF1 164512 120 - 1 UUUGCCAGUUUGAUGGCUUCGCCAGCUCCGCUAACUGCCACUAUCAGCGAAAUGAAAUUGAUUGGAAAAUGCCAAUGAAAAGAUUGCAUUGUGUAGCUAUUAAGCGGGGGAAUAGUUCCA ....((.(((((((((((.(((..((..((((.............))))........((.(((((......))))).))......))...))).))))))))))).))(((.....))). ( -31.02) >DroYak_CAF1 167264 120 - 1 UUUGCCAGUUUGAUGGCAACGCCAGCUUCGCUAACUGCCACUAUCAGCGAAAUGCAAUUGAUUGGAAAAUGCCAAUGAAAAGAUUGCAUUGUGUAGCUAUUAAGCGGGCGAAUAGUUCUU ((((((.((((((((((.((((.....(((((.............)))))(((((((((.(((((......))))).....))))))))))))).)))))))))).))))))........ ( -41.82) >consensus UUUGCCAGUUUGAUGGCUUCGCCAGCUCCGCUAACUGCCACUAUCAGCGAAAUGAAAUUGAUUGGAAAAUGCCAAUGAAAAGAUUGCAUUGUGCAGCUAUUAAGCGGGGGAAUAGUUCUU (((.((.(((((((((((.(((..((..((((.............))))........((.(((((......))))).))......))...))).))))))))))).)).)))........ (-28.34 = -28.94 + 0.60)



| Location | 11,434,875 – 11,434,995 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.50 |
| Mean single sequence MFE | -37.70 |
| Consensus MFE | -35.10 |
| Energy contribution | -35.34 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.83 |
| Structure conservation index | 0.93 |
| SVM decision value | 4.05 |
| SVM RNA-class probability | 0.999776 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11434875 120 + 27905053 UUUCAUUGGCAUUUUCCAAUCAAUUUCAUUUCGCUGAUAGUGGCAGUUAGCGAAGCUGGCGAAGCCAUCAAACUGGCAAACUAUCAGCCACCCAAGCGGCAAAUCCGCAGCCAGCAUAUG .....(((((......................((((((((((.(((((.....))))).)...((((......))))..))))))))).......((((.....)))).)))))...... ( -38.20) >DroSec_CAF1 174979 120 + 1 UUUCAUUGGCAUUUUCCAAUCAAUUUCAUUUCGCUGAUAGUGGCAGUUAGCGGAGCUGGCGAAGCCAUCAAACUGGCAAACUAUCAGCCACCCAAGCGGCAAAUCCGCAGCCAGCAUAUG .....(((((......................((((((((((.(((((.....))))).)...((((......))))..))))))))).......((((.....)))).)))))...... ( -38.30) >DroSim_CAF1 169190 120 + 1 UUUCAUUGGCAUUUUCCAAUCAAUUUCAUUUCGCUGAUAGUGGCAGUUAGCGUAGCUGGAGAAGCCAUCAAACUGGCAAACUAUCAGCCACCCAAGCGGCAAAUCCGCAGCCAGCAUAUG .....(((((......................(((((((((..((((((...)))))).....((((......))))..))))))))).......((((.....)))).)))))...... ( -36.10) >DroEre_CAF1 164552 120 + 1 UUUCAUUGGCAUUUUCCAAUCAAUUUCAUUUCGCUGAUAGUGGCAGUUAGCGGAGCUGGCGAAGCCAUCAAACUGGCAAACUAUCAACCACCCAAGCGGCAAAUCCGCAGCCAGCAUAUG ....(((((......))))).........(((((((((.......)))))))))((((((...((((......))))..................((((.....)))).))))))..... ( -36.90) >DroYak_CAF1 167304 120 + 1 UUUCAUUGGCAUUUUCCAAUCAAUUGCAUUUCGCUGAUAGUGGCAGUUAGCGAAGCUGGCGUUGCCAUCAAACUGGCAAACUAUCAACCACCCAAGCGGCAAAUCCGCAGCCAGCAUAUG ....(((((......))))).........(((((((((.......)))))))))((((((.((((((......))))))................((((.....)))).))))))..... ( -39.00) >consensus UUUCAUUGGCAUUUUCCAAUCAAUUUCAUUUCGCUGAUAGUGGCAGUUAGCGAAGCUGGCGAAGCCAUCAAACUGGCAAACUAUCAGCCACCCAAGCGGCAAAUCCGCAGCCAGCAUAUG ....(((((......))))).........(((((((((.......)))))))))((((((...((((......))))..................((((.....)))).))))))..... (-35.10 = -35.34 + 0.24)

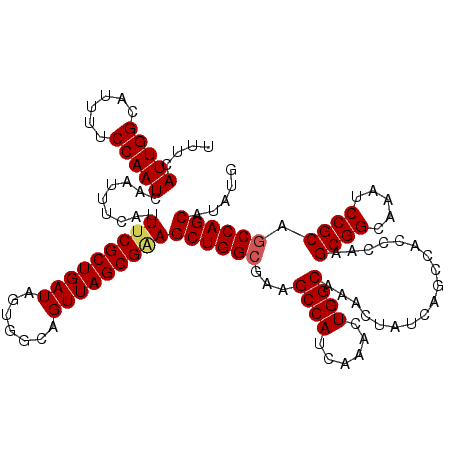

| Location | 11,434,915 – 11,435,017 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.85 |
| Mean single sequence MFE | -37.18 |
| Consensus MFE | -33.06 |
| Energy contribution | -33.10 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.40 |
| SVM RNA-class probability | 0.950787 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11434915 102 + 27905053 UGGCAGUUAGCGAAGCUGGCGAAGCCAUCAAACUGGCAAACUAUCAGCCACCCAAGCGGCAAAUCCGCAGCCAGCAUAUGCGACUGUUCCCGGUAACGGC---------GA--------- ..((((((.(((..((((((...((((......))))..................((((.....)))).))))))...)))))))))(((((....))).---------))--------- ( -36.80) >DroSec_CAF1 175019 102 + 1 UGGCAGUUAGCGGAGCUGGCGAAGCCAUCAAACUGGCAAACUAUCAGCCACCCAAGCGGCAAAUCCGCAGCCAGCAUAUGCGACUGUGCCCGGCGAUGGC---------GA--------- ..((((((.(((..((((((...((((......))))..................((((.....)))).))))))...)))))))))(((.......)))---------..--------- ( -37.70) >DroSim_CAF1 169230 102 + 1 UGGCAGUUAGCGUAGCUGGAGAAGCCAUCAAACUGGCAAACUAUCAGCCACCCAAGCGGCAAAUCCGCAGCCAGCAUAUGCGACUGUUCCCGGUGAUGGC---------GA--------- ..((((((.((((((((((....((((......))))..................((((.....))))..))))).)))))))))))(((((....))).---------))--------- ( -34.30) >DroEre_CAF1 164592 111 + 1 UGGCAGUUAGCGGAGCUGGCGAAGCCAUCAAACUGGCAAACUAUCAACCACCCAAGCGGCAAAUCCGCAGCCAGCAUAUGCGACUGUUCCCGCUGAUGGC---------GAAAAUGGUGA ..((.((((((((.((((((...((((......))))..................((((.....)))).))))))......((....)))))))))).))---------........... ( -38.80) >DroYak_CAF1 167344 120 + 1 UGGCAGUUAGCGAAGCUGGCGUUGCCAUCAAACUGGCAAACUAUCAACCACCCAAGCGGCAAAUCCGCAGCCAGCAUAUGCGACUGUUCCCGGUGAUGGCAAAAUUGGUGAAAAUGGUGA ..((((((.(((..((((((.((((((......))))))................((((.....)))).))))))...)))))))))(((((((.........))))).))......... ( -38.30) >consensus UGGCAGUUAGCGAAGCUGGCGAAGCCAUCAAACUGGCAAACUAUCAGCCACCCAAGCGGCAAAUCCGCAGCCAGCAUAUGCGACUGUUCCCGGUGAUGGC_________GA_________ ..((((((.(((..((((((...((((......))))..................((((.....)))).))))))...)))))))))..(((....)))..................... (-33.06 = -33.10 + 0.04)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:25:39 2006