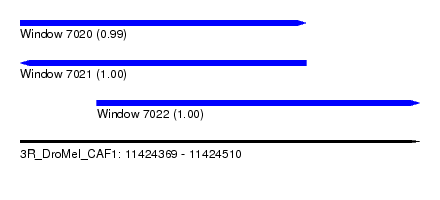
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 11,424,369 – 11,424,510 |
| Length | 141 |
| Max. P | 0.999624 |

| Location | 11,424,369 – 11,424,470 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.15 |
| Mean single sequence MFE | -25.92 |
| Consensus MFE | -22.18 |
| Energy contribution | -22.38 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.86 |
| SVM decision value | 2.37 |
| SVM RNA-class probability | 0.993082 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11424369 101 + 27905053 -------------AAGCACCCAUAUUCGCCCAGUUGGGCGCACAUUUUUGGCAUUUUUAUGACUCAUUGAGUCGAUUACUAUAGUGCUUGGCUAUCUUUAUAUAUAC------AUAUAUA -------------((((((........((((....))))((.((....)))).......((((((...)))))).........)))))).........((((((...------.)))))) ( -23.50) >DroSec_CAF1 171908 97 + 1 -------------AAGCACCCAUAUUCGCCCAGUUGGGCGCACAUUUUUGGCAUUUUUAUGACUCAUUGAGUCGAUUACUAAAGUGCUUGGCUAUCUUUGUAUAU----------AUAUA -------------.(((.........(((((....))))).........((((((((..((((((...))))))......))))))))..)))............----------..... ( -23.20) >DroSim_CAF1 166124 110 + 1 UGGCGGGAUCCCAAAGCACCCAUAUUCGCCCAUUUGGGCGCACAUUUUUGGCAUUUUUAUGACUCAUUGAGUCGAUUACUAUAGUGCUUGGCUAUCUUUGUAUAU----------AUAUA ((((((....)).((((((........((((....))))((.((....)))).......((((((...)))))).........)))))).))))...........----------..... ( -29.60) >DroEre_CAF1 161613 93 + 1 -------------AAGCACCCAUAUUCGCCCAGUUGGGCGCACAUUUUUGGCAUUUUUAUGACUCAUUGAGUCGAUUACUAUAGUGCUUGGCUAUCUUUAUA--------------UAUA -------------((((((........((((....))))((.((....)))).......((((((...)))))).........)))))).............--------------.... ( -23.00) >DroYak_CAF1 164185 107 + 1 -------------AAGCCCGCAUAUUCGCCCAGUUGGGCGCACAUUUUUGGCAUUUUUAUGACUCAUUGAGUCGAUUACUAUAGUGCUUGGCUAUCUUUAUAUAUACUCGUAUAUAUAUA -------------.((((.((((....((((....))))((.((....)))).......((((((...)))))).........))))..)))).....((((((((....)))))))).. ( -30.30) >consensus _____________AAGCACCCAUAUUCGCCCAGUUGGGCGCACAUUUUUGGCAUUUUUAUGACUCAUUGAGUCGAUUACUAUAGUGCUUGGCUAUCUUUAUAUAU__________AUAUA .............((((((........((((....))))((.((....)))).......((((((...)))))).........))))))............................... (-22.18 = -22.38 + 0.20)

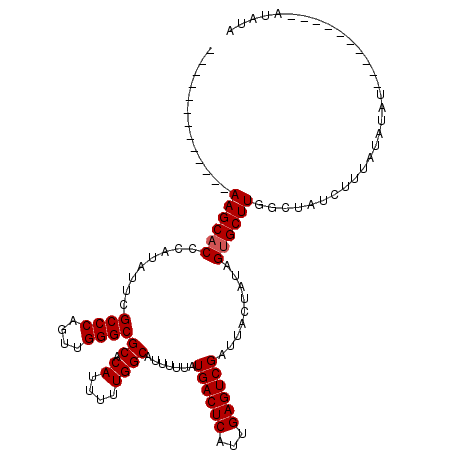
| Location | 11,424,369 – 11,424,470 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.15 |
| Mean single sequence MFE | -27.74 |
| Consensus MFE | -23.96 |
| Energy contribution | -24.36 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.28 |
| Structure conservation index | 0.86 |
| SVM decision value | 3.80 |
| SVM RNA-class probability | 0.999624 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11424369 101 - 27905053 UAUAUAU------GUAUAUAUAAAGAUAGCCAAGCACUAUAGUAAUCGACUCAAUGAGUCAUAAAAAUGCCAAAAAUGUGCGCCCAACUGGGCGAAUAUGGGUGCUU------------- ((((((.------...)))))).........(((((((...(((.(((((((...)))))........((((....)).))((((....)))))).))).)))))))------------- ( -27.20) >DroSec_CAF1 171908 97 - 1 UAUAU----------AUAUACAAAGAUAGCCAAGCACUUUAGUAAUCGACUCAAUGAGUCAUAAAAAUGCCAAAAAUGUGCGCCCAACUGGGCGAAUAUGGGUGCUU------------- .....----------................((((((((........(((((...)))))...............((((.(((((....))))).))))))))))))------------- ( -25.20) >DroSim_CAF1 166124 110 - 1 UAUAU----------AUAUACAAAGAUAGCCAAGCACUAUAGUAAUCGACUCAAUGAGUCAUAAAAAUGCCAAAAAUGUGCGCCCAAAUGGGCGAAUAUGGGUGCUUUGGGAUCCCGCCA .....----------.........(((..(((((((((...(((.(((((((...)))))........((((....)).))((((....)))))).))).))))).)))).)))...... ( -30.80) >DroEre_CAF1 161613 93 - 1 UAUA--------------UAUAAAGAUAGCCAAGCACUAUAGUAAUCGACUCAAUGAGUCAUAAAAAUGCCAAAAAUGUGCGCCCAACUGGGCGAAUAUGGGUGCUU------------- ....--------------.............(((((((...(((.(((((((...)))))........((((....)).))((((....)))))).))).)))))))------------- ( -26.10) >DroYak_CAF1 164185 107 - 1 UAUAUAUAUACGAGUAUAUAUAAAGAUAGCCAAGCACUAUAGUAAUCGACUCAAUGAGUCAUAAAAAUGCCAAAAAUGUGCGCCCAACUGGGCGAAUAUGCGGGCUU------------- (((((((((....))))))))).....((((..((............(((((...)))))...............((((.(((((....))))).)))))).)))).------------- ( -29.40) >consensus UAUAU__________AUAUAUAAAGAUAGCCAAGCACUAUAGUAAUCGACUCAAUGAGUCAUAAAAAUGCCAAAAAUGUGCGCCCAACUGGGCGAAUAUGGGUGCUU_____________ ...............................(((((((...(((.(((((((...)))))........((((....)).))((((....)))))).))).)))))))............. (-23.96 = -24.36 + 0.40)



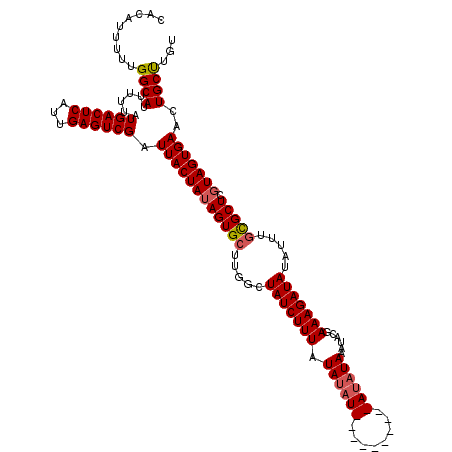
| Location | 11,424,396 – 11,424,510 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.58 |
| Mean single sequence MFE | -28.60 |
| Consensus MFE | -24.02 |
| Energy contribution | -24.62 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.85 |
| Structure conservation index | 0.84 |
| SVM decision value | 3.21 |
| SVM RNA-class probability | 0.998745 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11424396 114 + 27905053 CACAUUUUUGGCAUUUUUAUGACUCAUUGAGUCGAUUACUAUAGUGCUUGGCUAUCUUUAUAUAUAC------AUAUAUAAAAACGAAAGAUAUAUUUGCGCUCGUAGUGAAAUGCUUGU .........((((((....((((((...)))))).((((((((((((..(..(((((((.(((((..------..)))))......)))))))..)..))))).)))))))))))))... ( -27.50) >DroSec_CAF1 171935 110 + 1 CACAUUUUUGGCAUUUUUAUGACUCAUUGAGUCGAUUACUAAAGUGCUUGGCUAUCUUUGUAUAU----------AUAUAAAUACGAAAGAUAUAUUUGCGCUCGUAGUGAACUGCUAGU .......((((((......((((((...)))))).((((((.(((((..(..(((((((((((..----------......)))).)))))))..)..)))))..))))))..)))))). ( -29.30) >DroSim_CAF1 166164 110 + 1 CACAUUUUUGGCAUUUUUAUGACUCAUUGAGUCGAUUACUAUAGUGCUUGGCUAUCUUUGUAUAU----------AUAUAAAUACGAAAGAUAUAUUUGCGCUCGUAGUGAACUGCUAGU .......((((((......((((((...)))))).((((((((((((..(..(((((((((((..----------......)))).)))))))..)..))))).)))))))..)))))). ( -31.00) >DroEre_CAF1 161640 106 + 1 CACAUUUUUGGCAUUUUUAUGACUCAUUGAGUCGAUUACUAUAGUGCUUGGCUAUCUUUAUA--------------UAUAUGUACGAAAGAUAUAUUUGUGCUCGUAGUGAACUGCCUGA .........((((......((((((...)))))).(((((((((..(..(..(((((((.((--------------(....)))..)))))))..)..)..)).)))))))..))))... ( -25.80) >DroYak_CAF1 164212 120 + 1 CACAUUUUUGGCAUUUUUAUGACUCAUUGAGUCGAUUACUAUAGUGCUUGGCUAUCUUUAUAUAUACUCGUAUAUAUAUAUGUACGAAAGAUAUAUUUUCGCUCGUAGUGAACUGCCUGA .........((((......((((((...)))))).(((((((((((......((((((.........((((((((....))))))))))))))......)))).)))))))..))))... ( -29.40) >consensus CACAUUUUUGGCAUUUUUAUGACUCAUUGAGUCGAUUACUAUAGUGCUUGGCUAUCUUUAUAUAU__________AUAUAAAUACGAAAGAUAUAUUUGCGCUCGUAGUGAACUGCUUGU .........((((......((((((...)))))).((((((((((((.....(((((((.((((((........))))))......))))))).....))))).)))))))..))))... (-24.02 = -24.62 + 0.60)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:25:31 2006