
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 11,424,027 – 11,424,267 |
| Length | 240 |
| Max. P | 0.997965 |

| Location | 11,424,027 – 11,424,147 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.17 |
| Mean single sequence MFE | -37.00 |
| Consensus MFE | -33.38 |
| Energy contribution | -34.22 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.90 |
| SVM decision value | 2.97 |
| SVM RNA-class probability | 0.997965 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11424027 120 + 27905053 CGAGUGUUUGCUCUGCCAACUUCGGCCAGCCAGGAUAUUCCCUUAAUCCUGCCACUUGAUCACCAGCUGCCGGCGCGUUUCAAGUCCCCAAUCGACAAUCCUUUGCGCCAUAGGUAGUCU ((((((...(((..(((......))).)))((((((.........)))))).)))))).......((((((((((((......(((.......))).......))))))...)))))).. ( -36.72) >DroSec_CAF1 171566 120 + 1 CGAGUGUUUGCUCUGCCAACUCCGGCCAGCCAGGAUAUUCCCUUAAUCCUGCCACUUGAUCACCAGCUGCCGGCGCGUUUCAAGUCCCCAAUCGACAAUCCUUUGCGCCAUAGGUAGUCU ((((((...(((..(((......))).)))((((((.........)))))).)))))).......((((((((((((......(((.......))).......))))))...)))))).. ( -36.72) >DroSim_CAF1 165404 120 + 1 CGAGUGUUUGCUCCGCCAACUCCGGCCAGGCCGGAUAUUCCCUUAAUCCUGCCACUUGAUCACCAGCUGCCGGCGCGUUUCAAGUCCCCAAUCGACAAUCCUUUGCGCCAUAGGUAGUCU (((((.........(((......)))..(((.((((.........)))).)))))))).......((((((((((((......(((.......))).......))))))...)))))).. ( -35.32) >DroEre_CAF1 161270 120 + 1 CGAGUGUUUGCUCUGCCAACUCCGACCAGGCAGGAUAUUCCUUUAAUCCUGCCACUUGAUCACCAGCUGCCGGCGCGUUUCAAGUCCCCAAUCGACAAUCCUUCGCGCCAUAGGUAGUCU ((((((((.((...)).)))........((((((((.........))))))))))))).......((((((((((((......(((.......))).......))))))...)))))).. ( -38.62) >DroYak_CAF1 163842 120 + 1 CGAGUGUUUGCUCUACCAACUCCGGCCAGGCAGGAUAUUCCCUUAAUCCUGCCACUUGAUCACCAGCUGCCGGCGCGUUUCAAGUCCCCAAUCGACAAUCCUUUGCGCCAUAGGUAGUCU .((((....))))((((......(((..((((((((.........))))))))..(((.....)))..)))((((((......(((.......))).......))))))...)))).... ( -37.62) >consensus CGAGUGUUUGCUCUGCCAACUCCGGCCAGGCAGGAUAUUCCCUUAAUCCUGCCACUUGAUCACCAGCUGCCGGCGCGUUUCAAGUCCCCAAUCGACAAUCCUUUGCGCCAUAGGUAGUCU (((((.........(((......)))..((((((((.........))))))))))))).......((((((((((((......(((.......))).......))))))...)))))).. (-33.38 = -34.22 + 0.84)

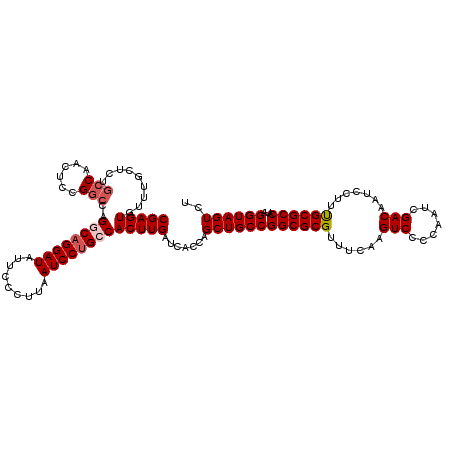

| Location | 11,424,067 – 11,424,187 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.17 |
| Mean single sequence MFE | -31.02 |
| Consensus MFE | -28.38 |
| Energy contribution | -28.62 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.91 |
| SVM decision value | 2.50 |
| SVM RNA-class probability | 0.994714 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11424067 120 + 27905053 CCUUAAUCCUGCCACUUGAUCACCAGCUGCCGGCGCGUUUCAAGUCCCCAAUCGACAAUCCUUUGCGCCAUAGGUAGUCUCGCCAGUCUAUUAAGUGGCGCUCCCCAUAAUUCACAACAC ..........((((((((((.((.(((((((((((((......(((.......))).......))))))...))))).)).....))..))))))))))..................... ( -30.52) >DroSec_CAF1 171606 120 + 1 CCUUAAUCCUGCCACUUGAUCACCAGCUGCCGGCGCGUUUCAAGUCCCCAAUCGACAAUCCUUUGCGCCAUAGGUAGUCUCGCCAGUCUAUUAAGUGGCGCUCCCCAUAAUUCACAACAC ..........((((((((((.((.(((((((((((((......(((.......))).......))))))...))))).)).....))..))))))))))..................... ( -30.52) >DroSim_CAF1 165444 120 + 1 CCUUAAUCCUGCCACUUGAUCACCAGCUGCCGGCGCGUUUCAAGUCCCCAAUCGACAAUCCUUUGCGCCAUAGGUAGUCUCGCCAGUCUAUUAAGUGGCGCUCCCCAUAAUUCACAACAC ..........((((((((((.((.(((((((((((((......(((.......))).......))))))...))))).)).....))..))))))))))..................... ( -30.52) >DroEre_CAF1 161310 120 + 1 CUUUAAUCCUGCCACUUGAUCACCAGCUGCCGGCGCGUUUCAAGUCCCCAAUCGACAAUCCUUCGCGCCAUAGGUAGUCUCGCCGGUCUAUUAAGUGGGGCUGGCCAUAAUUCACAACAC ...........(((((((((.((((((((((((((((......(((.......))).......))))))...))))).))....)))..)))))))))((....)).............. ( -34.32) >DroYak_CAF1 163882 120 + 1 CCUUAAUCCUGCCACUUGAUCACCAGCUGCCGGCGCGUUUCAAGUCCCCAAUCGACAAUCCUUUGCGCCAUAGGUAGUCUCGCCAGUCUAUUAAGUGGGCCUGGCCAUAAUUCACAACAC ................(((......((((((((((((......(((.......))).......))))))...))))))...((((((((((...))))).)))))......)))...... ( -29.22) >consensus CCUUAAUCCUGCCACUUGAUCACCAGCUGCCGGCGCGUUUCAAGUCCCCAAUCGACAAUCCUUUGCGCCAUAGGUAGUCUCGCCAGUCUAUUAAGUGGCGCUCCCCAUAAUUCACAACAC ..........((((((((((.((.(((((((((((((......(((.......))).......))))))...))))).)).....))..))))))))))..................... (-28.38 = -28.62 + 0.24)

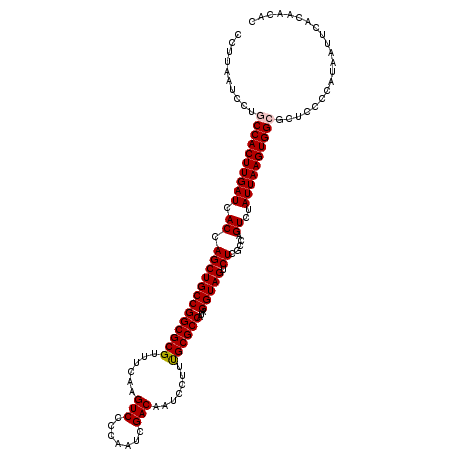
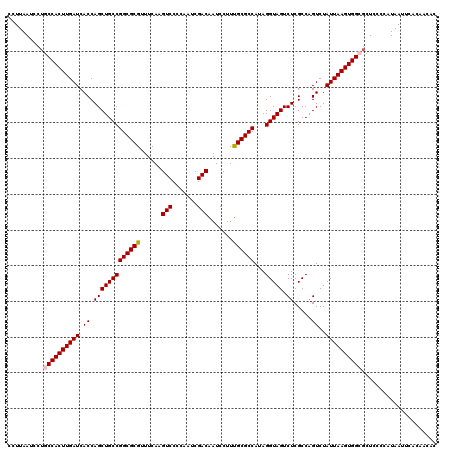
| Location | 11,424,147 – 11,424,267 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.58 |
| Mean single sequence MFE | -39.90 |
| Consensus MFE | -33.52 |
| Energy contribution | -33.76 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.58 |
| SVM RNA-class probability | 0.965260 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11424147 120 + 27905053 CGCCAGUCUAUUAAGUGGCGCUCCCCAUAAUUCACAACACUUUCGGUCCAUCAAGGACCAAAGGGAUGCAGAGCGUCCAGCUGAGGCCAGCAUAAGGCGCAUUUCGCAAACAAAAAGCGA ((((...........((((((((..(((...........((((.(((((.....))))).)))).)))..))))).)))((((....))))....))))....((((.........)))) ( -38.60) >DroSec_CAF1 171686 120 + 1 CGCCAGUCUAUUAAGUGGCGCUCCCCAUAAUUCACAACACUUUCGGUCCAUCAAGGACGAAAGGGAUGCAGAGCGUCCAGCUGAGGCCAGCAUAAGGCGCAUUUCGCAAACAAAAAGCGA ((((...........((((((((..(((...........((((((.(((.....)))))))))..)))..))))).)))((((....))))....))))....((((.........)))) ( -37.62) >DroSim_CAF1 165524 120 + 1 CGCCAGUCUAUUAAGUGGCGCUCCCCAUAAUUCACAACACUUUCGGUCCAUCAAGGACGAAAGGGAUGCAGAGCGUCCAGCUGAGGCCAGCAUAGGGCGCAUUUCGCAAACAAAAAGCGA .((((.(......).))))((.(((..............((((((.(((.....)))))))))((((((...)))))).((((....))))...))).))...((((.........)))) ( -40.80) >DroEre_CAF1 161390 120 + 1 CGCCGGUCUAUUAAGUGGGGCUGGCCAUAAUUCACAACACUUUCGGUCCAUCAAGGACGAUGGGGAUGCAGAGCGUCCAGCUGAGGCCAGCAUAAGGCGCAUUUCGCAAACACAAAGCGA ((((..(((((...)))))(((((((.............((.(((.(((.....)))))).))((((((...))))))......)))))))....))))....((((.........)))) ( -41.60) >DroYak_CAF1 163962 120 + 1 CGCCAGUCUAUUAAGUGGGCCUGGCCAUAAUUCACAACACUUUCGGUCCAUCAAGGACGAGGGGGCUGCAGAGCGUCCAGCUGGGGCCAGCAUAAGGCGCAUUUCGCAAACACAAUGCGA ((((.((((((...))))))((((((.............((((((.(((.....)))))))))(((((.........)))))..)))))).....))))....(((((.......))))) ( -40.90) >consensus CGCCAGUCUAUUAAGUGGCGCUCCCCAUAAUUCACAACACUUUCGGUCCAUCAAGGACGAAAGGGAUGCAGAGCGUCCAGCUGAGGCCAGCAUAAGGCGCAUUUCGCAAACAAAAAGCGA ((((..........((((......))))...........((((((.(((.....)))))))))((((((...)))))).((((....))))....))))....((((.........)))) (-33.52 = -33.76 + 0.24)


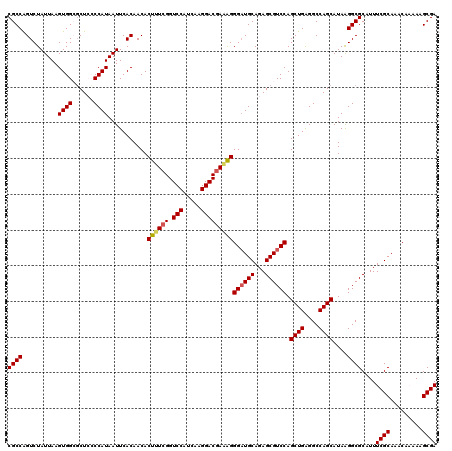
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:25:28 2006