
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 11,423,115 – 11,423,272 |
| Length | 157 |
| Max. P | 0.997876 |

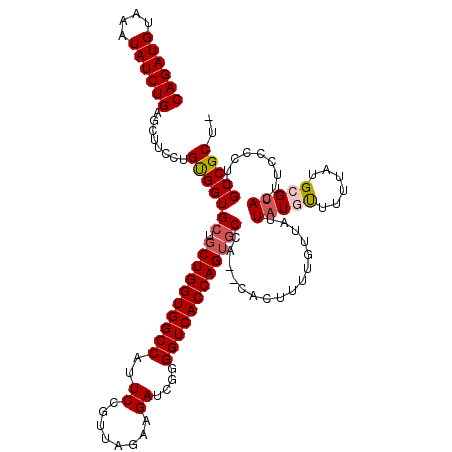
| Location | 11,423,115 – 11,423,232 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.11 |
| Mean single sequence MFE | -35.30 |
| Consensus MFE | -30.06 |
| Energy contribution | -30.66 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.43 |
| SVM RNA-class probability | 0.953289 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11423115 117 - 27905053 CAGAUGUAAAUAUCUGAGCUUCCUGUGGUGCUGCUGGUGGCCAUUCUGUUAGAAGAUCGGGGUCACCAGUGCCA--CACUUUUGUUAUUAUGUUUUUAUGCGAACUUCCCCUUGCCGCU- ((((((....))))))(((....((((((...((((((((((..(((......)))....))))))))))))))--)).....................((((........)))).)))- ( -36.90) >DroSec_CAF1 170641 117 - 1 CAGAUGUAAAUAUCUGAGCUUCCUGUGGUGCUGCUGGUGGCCAUUCAGUUAGAAGAUCGGGGUCACCAGUGCCA--CACUUUUGUUAUUAUGCUUUUAUGCGUACUUCCCCUUGCCGCU- ((((((....))))))(((....((((((...((((((((((..((........))....))))))))))))))--))..........(((((......)))))............)))- ( -37.20) >DroSim_CAF1 164501 117 - 1 CAGAUGUAAAUAUCUGAGCUUCCUGUGGUGCUGCUGGUGGCCAUUCUGUUAGAAGAUCGGGGUCACCAGGACCA--CACUUUUGUUAUUAUGCUUUUAUGCGUACUUCCCCUUGCCGCU- ((((((....))))))(((....((((((....(((((((((..(((......)))....))))))))).))))--))..........(((((......)))))............)))- ( -36.80) >DroEre_CAF1 160350 119 - 1 CAGAUGUAAAUAUCUGA-UUUCCUGUGGUGCUGCUGGUGGCCAUUCCGUUAGAAGAUCGAGGUCACCAGUGCCACACACUUUUGUUAUUAUCUUUUUAUGCGUACUUCCCCUUGCCGCAC ((((((....)))))).-.....((((((...((((((((((..((.(((....))).))))))))))))))))))......................((((((........)).)))). ( -32.30) >DroYak_CAF1 162937 119 - 1 CAGAUGUAAAUAUCUGG-UUUCCUGCGGUGCUGCUGGUGGCCAUUCCGUUAGAAGAUCGGGGUCACCAGUGCCACACACUUUUGUUAUUAUCUUUUUAUGCGUACUUCCCCCUGCCGCUC ((((((....)))))).-......(((((((.((((((((((...(((.........))))))))))))))).(((......)))............................))))).. ( -33.30) >consensus CAGAUGUAAAUAUCUGAGCUUCCUGUGGUGCUGCUGGUGGCCAUUCCGUUAGAAGAUCGGGGUCACCAGUGCCA__CACUUUUGUUAUUAUGUUUUUAUGCGUACUUCCCCUUGCCGCU_ ((((((....))))))........(((((((.((((((((((..((........))....))))))))))))................(((((......))))).........))))).. (-30.06 = -30.66 + 0.60)


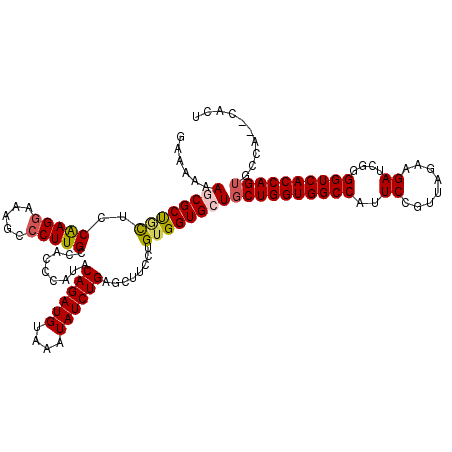
| Location | 11,423,154 – 11,423,272 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.79 |
| Mean single sequence MFE | -40.08 |
| Consensus MFE | -35.02 |
| Energy contribution | -35.34 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.87 |
| SVM decision value | 2.95 |
| SVM RNA-class probability | 0.997876 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11423154 118 - 27905053 GAAAAAAGCGCUGCUCCAAGGAAAGCCCUUGCCACCCAUACAGAUGUAAAUAUCUGAGCUUCCUGUGGUGCUGCUGGUGGCCAUUCUGUUAGAAGAUCGGGGUCACCAGUGCCA--CACU .....((((.......(((((.....))))).........((((((....)))))).))))..((((((...((((((((((..(((......)))....))))))))))))))--)).. ( -41.90) >DroSec_CAF1 170680 118 - 1 GAAAAAAUCGCUGCUCCAAGGAAAGCCCUUGCCACCCAUACAGAUGUAAAUAUCUGAGCUUCCUGUGGUGCUGCUGGUGGCCAUUCAGUUAGAAGAUCGGGGUCACCAGUGCCA--CACU .........(((....(((((.....))))).........((((((....)))))))))....((((((...((((((((((..((........))....))))))))))))))--)).. ( -39.10) >DroSim_CAF1 164540 118 - 1 GAAAAAAUCGCUGCUCCAAGGAAAGCCCUUGCCACCCAUACAGAUGUAAAUAUCUGAGCUUCCUGUGGUGCUGCUGGUGGCCAUUCUGUUAGAAGAUCGGGGUCACCAGGACCA--CACU .........(((....(((((.....))))).........((((((....)))))))))....((((((....(((((((((..(((......)))....))))))))).))))--)).. ( -38.70) >DroEre_CAF1 160390 119 - 1 GAAAAAAGCGCCACUCCAAGGAAAGUCCUUGCCACCCAUACAGAUGUAAAUAUCUGA-UUUCCUGUGGUGCUGCUGGUGGCCAUUCCGUUAGAAGAUCGAGGUCACCAGUGCCACACACU ......((((((((..(((((.....))))).........((((((....)))))).-......))))))))((((((((((..((.(((....))).)))))))))))).......... ( -42.30) >DroYak_CAF1 162977 119 - 1 GGAAAAAGCGCCAUUACGAGAAAAACCCUUGCCACCCAUACAGAUGUAAAUAUCUGG-UUUCCUGCGGUGCUGCUGGUGGCCAUUCCGUUAGAAGAUCGGGGUCACCAGUGCCACACACU ((....((((((....((((.......)))).........((((((....)))))).-........))))))((((((((((...(((.........))))))))))))).))....... ( -38.40) >consensus GAAAAAAGCGCUGCUCCAAGGAAAGCCCUUGCCACCCAUACAGAUGUAAAUAUCUGAGCUUCCUGUGGUGCUGCUGGUGGCCAUUCCGUUAGAAGAUCGGGGUCACCAGUGCCA__CACU ......((((((((..(((((.....))))).........((((((....))))))........))))))))((((((((((..((........))....)))))))))).......... (-35.02 = -35.34 + 0.32)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:25:23 2006