
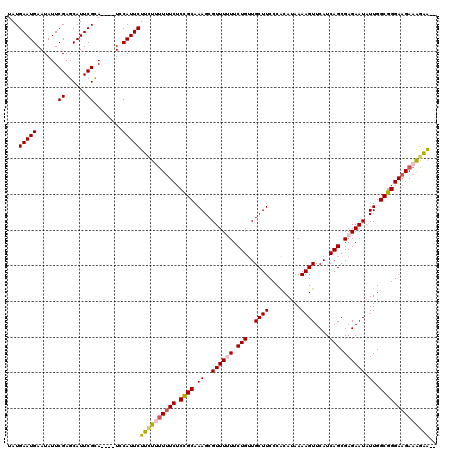
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 11,422,171 – 11,422,286 |
| Length | 115 |
| Max. P | 0.823452 |

| Location | 11,422,171 – 11,422,286 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.95 |
| Mean single sequence MFE | -28.02 |
| Consensus MFE | -22.36 |
| Energy contribution | -22.92 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.581854 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11422171 115 + 27905053 UAUGAAUGAAUAUUCGAGCAUUCGCA----UCCAUUCUUCUUUUUUCUCCGCAAAGCGUUUUUUCUGUUGCUUCCCACAUAAAAGUUCAUCAGCGAGAAUAUUGGCGGGAAGAAAAAA-G ...(((((.((...((......)).)----).)))))..(((((((((((((.((..((((((.(((..((((.........))))....))).)))))).)).))))..))))))))-) ( -23.70) >DroSec_CAF1 169646 116 + 1 UAUGAAUGAAUAUUCGAGCAUUCGUA----UCCAUUCUUCUUUUUUCUCCGCAAAGCGUUUUUUCUGAUGCUUCCCACAUAAAAGUUCAUCAGCGAGAAUAUUGGCGGGAAGAAAGAAGA ((((((((..........))))))))----.....((((((((((((.((((.((..((((((.(((((((((.........)))..)))))).)))))).)).)))))))))))))))) ( -36.60) >DroSim_CAF1 162472 115 + 1 UAUGAAUGAAUAUUCGAGCAUUCGCA----UCCAUUCUUCUUUUUUCUCCGCAAAGCGUUUUUUCUGUUGCUUCCCACAUAAAAGUUCAUCAGCGAGAAUAUUGGCGGGAAGAAAGAA-A ...(((((.((...((......)).)----).)))))((((((((((.((((.((..((((((.(((..((((.........))))....))).)))))).)).))))))))))))))-. ( -28.10) >DroEre_CAF1 157711 112 + 1 UAUGAAUGAAUAUUCGAGCAUUCGCA----UGCAUUCUUCU-CUUUCUCUGCAAAGCGUUU-UUCUGUUGCUUCCCGCAUAAAAGUUCAUCAGCGAGAAUAUUGGCGGGAAGAAAGGA-- ...(((((..(((.((......)).)----)))))))...(-(((((...(((..(((...-...))))))(((((((...((.((((........)))).)).))))))))))))).-- ( -25.90) >DroYak_CAF1 161903 116 + 1 UAUGAAUGAAUAUUCGAGCAUUCGCAUUCAUGCAUUCUUUU-UUCUCUCUGCAAAGCGUUU-UUCUGUUGCUUCCCGCAUAAAAGUUCAUCAGCGAGAAUAUUGGCGGGAAGAAGGAG-- ((((((((...((......))...)))))))).........-....((((....((((...-...)))).((((((((...((.((((........)))).)).))))))))..))))-- ( -25.80) >consensus UAUGAAUGAAUAUUCGAGCAUUCGCA____UCCAUUCUUCUUUUUUCUCCGCAAAGCGUUUUUUCUGUUGCUUCCCACAUAAAAGUUCAUCAGCGAGAAUAUUGGCGGGAAGAAAGAA__ ...(((((......((......))........)))))((((((((((.((((.((..((((((.(((..((((.........))))....))).)))))).)).)))))))))))))).. (-22.36 = -22.92 + 0.56)


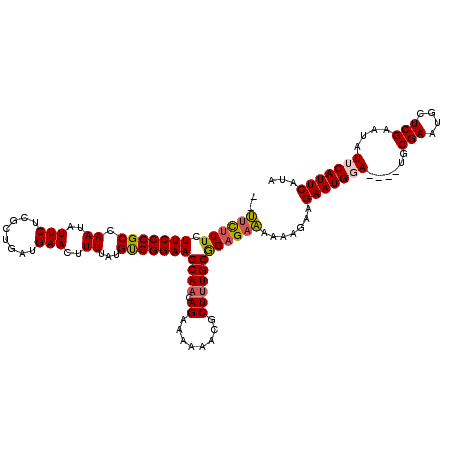
| Location | 11,422,171 – 11,422,286 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.95 |
| Mean single sequence MFE | -28.96 |
| Consensus MFE | -21.98 |
| Energy contribution | -22.02 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.823452 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11422171 115 - 27905053 C-UUUUUUCUUCCCGCCAAUAUUCUCGCUGAUGAACUUUUAUGUGGGAAGCAACAGAAAAAACGCUUUGCGGAGAAAAAAGAAGAAUGGA----UGCGAAUGCUCGAAUAUUCAUUCAUA (-((((((((((.........((((((((((.......))).)))))))((((.((........)))))))))))))))))..(((((((----(((((....)))..)))))))))... ( -31.60) >DroSec_CAF1 169646 116 - 1 UCUUCUUUCUUCCCGCCAAUAUUCUCGCUGAUGAACUUUUAUGUGGGAAGCAUCAGAAAAAACGCUUUGCGGAGAAAAAAGAAGAAUGGA----UACGAAUGCUCGAAUAUUCAUUCAUA ..((((((.(((((((...........((((((..(((((.....)))))))))))............)))).))).))))))(((((((----(((((....)))..)))))))))... ( -31.80) >DroSim_CAF1 162472 115 - 1 U-UUCUUUCUUCCCGCCAAUAUUCUCGCUGAUGAACUUUUAUGUGGGAAGCAACAGAAAAAACGCUUUGCGGAGAAAAAAGAAGAAUGGA----UGCGAAUGCUCGAAUAUUCAUUCAUA .-((((((.(((((((.....((((((((((.......))).)))))))((............))...)))).))).))))))(((((((----(((((....)))..)))))))))... ( -30.60) >DroEre_CAF1 157711 112 - 1 --UCCUUUCUUCCCGCCAAUAUUCUCGCUGAUGAACUUUUAUGCGGGAAGCAACAGAA-AAACGCUUUGCAGAGAAAG-AGAAGAAUGCA----UGCGAAUGCUCGAAUAUUCAUUCAUA --((((((((((((((.((..(((........)))..))...)))))))((((.((..-.....))))))...)))))-.)).(((((.(----(((((....)))..))).)))))... ( -25.90) >DroYak_CAF1 161903 116 - 1 --CUCCUUCUUCCCGCCAAUAUUCUCGCUGAUGAACUUUUAUGCGGGAAGCAACAGAA-AAACGCUUUGCAGAGAGAA-AAAAGAAUGCAUGAAUGCGAAUGCUCGAAUAUUCAUUCAUA --(((((..(((((((.((..(((........)))..))...)))))))((((.((..-.....)))))))).)))..-......(((.((((((((((....)))..))))))).))). ( -24.90) >consensus __UUCUUUCUUCCCGCCAAUAUUCUCGCUGAUGAACUUUUAUGUGGGAAGCAACAGAAAAAACGCUUUGCGGAGAAAAAAGAAGAAUGGA____UGCGAAUGCUCGAAUAUUCAUUCAUA ..((((((.(((((((.((..(((........)))..))...)))))))((((.((........)))))))))))).......(((((((......(((....)))....)))))))... (-21.98 = -22.02 + 0.04)

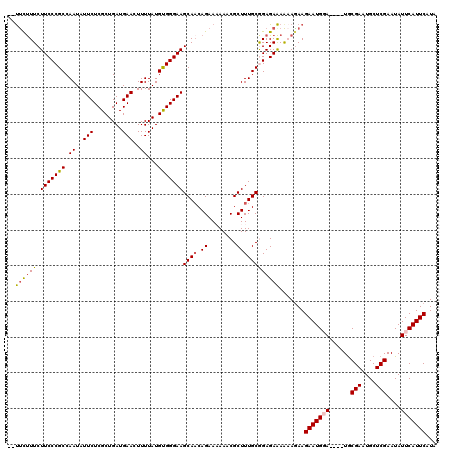
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:25:20 2006