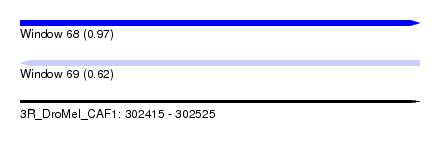
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 302,415 – 302,525 |
| Length | 110 |
| Max. P | 0.965414 |

| Location | 302,415 – 302,525 |
|---|---|
| Length | 110 |
| Sequences | 3 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 84.48 |
| Mean single sequence MFE | -30.90 |
| Consensus MFE | -20.01 |
| Energy contribution | -20.57 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.07 |
| Mean z-score | -3.83 |
| Structure conservation index | 0.65 |
| SVM decision value | 1.58 |
| SVM RNA-class probability | 0.965414 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
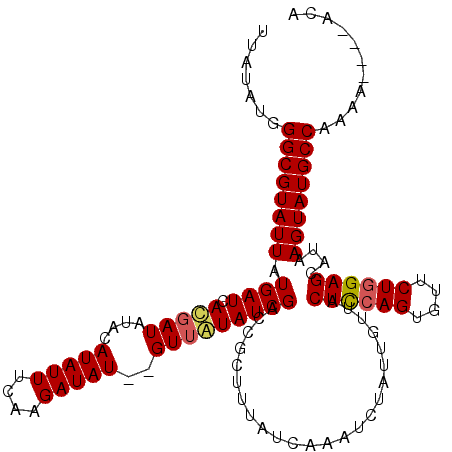
>3R_DroMel_CAF1 302415 110 + 27905053 UUAUAUUGGCGUAUUAUGAUCAUUAU----AUAUUUCAAGAUAUAGGUUAUAUCGACCGCUAUAUCAAAUCUUUUGUUACUUAAGGGCUCUGGAGCCUAAGUAUGCCAAAA----GCA .....((((((((((...........----.........((((((((((.....))))..))))))..................(((((....))))).))))))))))..----... ( -26.60) >DroSec_CAF1 37638 112 + 1 UCAUAUGGGCGUAUUAUGAUCACGAUAUACAUAUUUCAAGAUAU--GUUGUAUCGACCCCUUUAUCAAAUCUAUUGUUACUCCAGUGUUCUGGAGCAUAAGUAUGCCAAAA----ACA .......(((((((((((....(((((((((((((....)))))--).)))))))........................((((((....))))))))).))))))))....----... ( -32.20) >DroSim_CAF1 39354 116 + 1 UUAUAUGGGCGUAUUAUGAUCACGAUAUACAUAUUUCAAGAUAU--GUUAUAUCGACCGCUUUAUCAAAUAUAUUGGUACUCCAGUGUUCUGGAGCAUAAGUAUGCCAAAAAAAAAAA .......(((((((((((....(((((((((((((....)))))--).))))))).......((((((.....))))))((((((....))))))))).))))))))........... ( -33.90) >consensus UUAUAUGGGCGUAUUAUGAUCACGAUAUACAUAUUUCAAGAUAU__GUUAUAUCGACCGCUUUAUCAAAUCUAUUGUUACUCCAGUGUUCUGGAGCAUAAGUAUGCCAAAA____ACA .......((((((((.((((.(((((....(((((....)))))..)))))))))........................((((((....))))))....))))))))........... (-20.01 = -20.57 + 0.56)

| Location | 302,415 – 302,525 |
|---|---|
| Length | 110 |
| Sequences | 3 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 84.48 |
| Mean single sequence MFE | -25.80 |
| Consensus MFE | -16.02 |
| Energy contribution | -17.13 |
| Covariance contribution | 1.11 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.618704 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
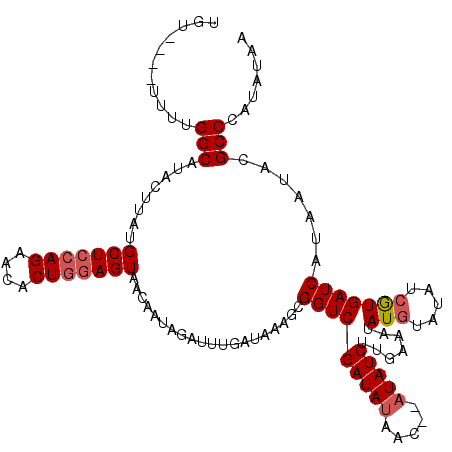
>3R_DroMel_CAF1 302415 110 - 27905053 UGC----UUUUGGCAUACUUAGGCUCCAGAGCCCUUAAGUAACAAAAGAUUUGAUAUAGCGGUCGAUAUAACCUAUAUCUUGAAAUAU----AUAAUGAUCAUAAUACGCCAAUAUAA ...----..(((((.((((((((((....))))..))))))......((((..(((((......((((((...))))))......)))----))...)))).......)))))..... ( -25.20) >DroSec_CAF1 37638 112 - 1 UGU----UUUUGGCAUACUUAUGCUCCAGAACACUGGAGUAACAAUAGAUUUGAUAAAGGGGUCGAUACAAC--AUAUCUUGAAAUAUGUAUAUCGUGAUCAUAAUACGCCCAUAUGA ...----....(((...(((.((((((((....)))))))).(((.....)))...)))((..(((((..((--((((......)))))).)))))...)).......)))....... ( -25.50) >DroSim_CAF1 39354 116 - 1 UUUUUUUUUUUGGCAUACUUAUGCUCCAGAACACUGGAGUACCAAUAUAUUUGAUAAAGCGGUCGAUAUAAC--AUAUCUUGAAAUAUGUAUAUCGUGAUCAUAAUACGCCCAUAUAA ...........(((.......((((((((....))))))))...................((((((((((.(--((((......)))))))))))..)))).......)))....... ( -26.70) >consensus UGU____UUUUGGCAUACUUAUGCUCCAGAACACUGGAGUAACAAUAGAUUUGAUAAAGCGGUCGAUAUAAC__AUAUCUUGAAAUAUGUAUAUCGUGAUCAUAAUACGCCCAUAUAA ...........(((........(((((((....)))))))....................(((((((((.....))))).......(((.....))))))).......)))....... (-16.02 = -17.13 + 1.11)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:31:10 2006