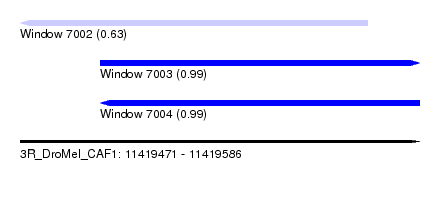
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 11,419,471 – 11,419,586 |
| Length | 115 |
| Max. P | 0.992066 |

| Location | 11,419,471 – 11,419,571 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.02 |
| Mean single sequence MFE | -30.04 |
| Consensus MFE | -21.24 |
| Energy contribution | -21.36 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.630563 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11419471 100 - 27905053 ---UUUUCGGCUGCCUGUAUUUGAACCAAGCACUUCAAAAGCAGCGUUAGAUUGCAUAAAAGCAAAUGCGGAAGUGGUAACUGCAACUGAAAGGGC-----------------GGUGGAA ---..(((.(((((((.((.(((.....(.((((((....(((((....).))))......((....)).)))))).).....))).))...))))-----------------))).))) ( -29.70) >DroSec_CAF1 167031 100 - 1 ---UUUUCGGCUGCCUGUAUUUGAACCAAGCACUUCAAAAGCAGCGUUAGAUUGCAUAAAAGCAAAUGCGGAAGUGGUAACUGCAGCUGAAAGGGC-----------------GGUGGGA ---(((((((((((((((.((((((........)))))).)))).((((..((((......))))..((....))..)))).)))))))))))...-----------------....... ( -30.70) >DroSim_CAF1 159849 100 - 1 ---UUUUCGGCUGCCUGUAUUUGAACCAAGCACUUCAAAAGCAGCGUUAGAUUGCAUAAAAGCAAAUGCGGAAGUGGUAACUGCAUCUGAAAGGGC-----------------GGUGGGA ---..(((.(((((((((.((((((........)))))).))....(((((((((......)))).(((((.........))))))))))..))))-----------------))).))) ( -28.80) >DroEre_CAF1 155007 120 - 1 CUUUUUUCGGCUGGCUGUAUUUGAAGCAAGCACUUCAAAAGCAGCGUUAGAUUGCAUAAAAGCAAAUGCGGAAGUGGUAACUGCAACUGAGGGGGCGGGUGGGGUGGUUAAGCGGUGGGA .....(((.(((.(((((.(((((((......))))))).)))))(((.(((..(......((....((....)).....((((..(....)..))))))...)..))).)))))).))) ( -35.30) >DroYak_CAF1 159162 93 - 1 ---GUUUCGACUGGCUGUAUUUGAAGCAAGCACUUCAAAAGCAGCGUUAGAUUGCAUAAAAGCAAAUGCGGAAGUGGUAACUGCAACUGAGGG------------------------GGA ---.(..(.....(((((.(((((((......))))))).))))).((((.((((......)))).(((((.........))))).)))))..------------------------).. ( -25.70) >consensus ___UUUUCGGCUGCCUGUAUUUGAACCAAGCACUUCAAAAGCAGCGUUAGAUUGCAUAAAAGCAAAUGCGGAAGUGGUAACUGCAACUGAAAGGGC_________________GGUGGGA ....((((((.(((((((.((((((........)))))).)))).((((..((((......))))..((....))..)))).))).))))))............................ (-21.24 = -21.36 + 0.12)

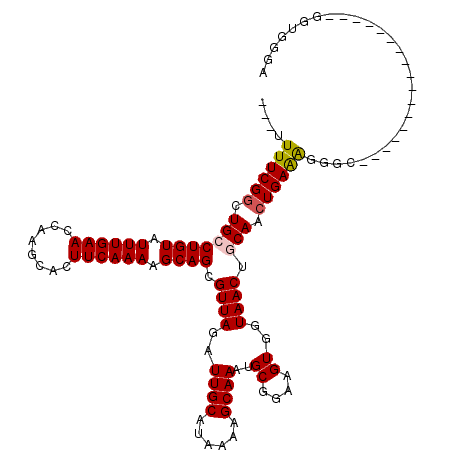
| Location | 11,419,494 – 11,419,586 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.09 |
| Mean single sequence MFE | -26.74 |
| Consensus MFE | -18.96 |
| Energy contribution | -19.80 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.05 |
| Mean z-score | -3.20 |
| Structure conservation index | 0.71 |
| SVM decision value | 2.30 |
| SVM RNA-class probability | 0.992066 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11419494 92 + 27905053 UUACCACUUCCGCAUUUGCUUUUAUGCAAUCUAACGCUGCUUUUGAAGUGCUUGGUUCAAAUACAGGCAGCCGAAAA----------------------------AAGCGGAAAUAAAAU .......((((((..((((......))))......((((((((((((.(....).))))))....))))))......----------------------------..))))))....... ( -25.70) >DroSec_CAF1 167054 92 + 1 UUACCACUUCCGCAUUUGCUUUUAUGCAAUCUAACGCUGCUUUUGAAGUGCUUGGUUCAAAUACAGGCAGCCGAAAA----------------------------AAGCGGAAAUAAAAU .......((((((..((((......))))......((((((((((((.(....).))))))....))))))......----------------------------..))))))....... ( -25.70) >DroSim_CAF1 159872 92 + 1 UUACCACUUCCGCAUUUGCUUUUAUGCAAUCUAACGCUGCUUUUGAAGUGCUUGGUUCAAAUACAGGCAGCCGAAAA----------------------------AAGCGGAAAUAAAAU .......((((((..((((......))))......((((((((((((.(....).))))))....))))))......----------------------------..))))))....... ( -25.70) >DroEre_CAF1 155047 120 + 1 UUACCACUUCCGCAUUUGCUUUUAUGCAAUCUAACGCUGCUUUUGAAGUGCUUGCUUCAAAUACAGCCAGCCGAAAAAAGCCAAAAAGGGCGAAAAAGGGGGAAAAAGCGGAAAUAAAAU .......((((((..((((......))))(((..(((((..((((((((....))))))))..))))............(((......)))......)..)))....))))))....... ( -36.10) >DroYak_CAF1 159178 100 + 1 UUACCACUUCCGCAUUUGCUUUUAUGCAAUCUAACGCUGCUUUUGAAGUGCUUGCUUCAAAUACAGCCAGUCGAAAC-------------CGAAAA-------AAAACCAAAAAAAAAGU .......(((.((..((((......))))......((((..((((((((....))))))))..))))..)).)))..-------------......-------................. ( -20.50) >consensus UUACCACUUCCGCAUUUGCUUUUAUGCAAUCUAACGCUGCUUUUGAAGUGCUUGGUUCAAAUACAGGCAGCCGAAAA____________________________AAGCGGAAAUAAAAU .......((((((..((((......))))......((((((((((((........))))))....))))))....................................))))))....... (-18.96 = -19.80 + 0.84)

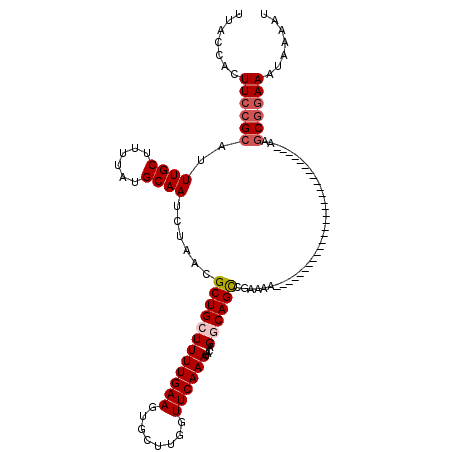

| Location | 11,419,494 – 11,419,586 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.09 |
| Mean single sequence MFE | -27.46 |
| Consensus MFE | -23.44 |
| Energy contribution | -23.36 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.85 |
| SVM decision value | 2.11 |
| SVM RNA-class probability | 0.988272 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11419494 92 - 27905053 AUUUUAUUUCCGCUU----------------------------UUUUCGGCUGCCUGUAUUUGAACCAAGCACUUCAAAAGCAGCGUUAGAUUGCAUAAAAGCAAAUGCGGAAGUGGUAA ...((((((((((..----------------------------......(((((.....((((((........)))))).)))))......((((......))))..))))))))))... ( -25.70) >DroSec_CAF1 167054 92 - 1 AUUUUAUUUCCGCUU----------------------------UUUUCGGCUGCCUGUAUUUGAACCAAGCACUUCAAAAGCAGCGUUAGAUUGCAUAAAAGCAAAUGCGGAAGUGGUAA ...((((((((((..----------------------------......(((((.....((((((........)))))).)))))......((((......))))..))))))))))... ( -25.70) >DroSim_CAF1 159872 92 - 1 AUUUUAUUUCCGCUU----------------------------UUUUCGGCUGCCUGUAUUUGAACCAAGCACUUCAAAAGCAGCGUUAGAUUGCAUAAAAGCAAAUGCGGAAGUGGUAA ...((((((((((..----------------------------......(((((.....((((((........)))))).)))))......((((......))))..))))))))))... ( -25.70) >DroEre_CAF1 155047 120 - 1 AUUUUAUUUCCGCUUUUUCCCCCUUUUUCGCCCUUUUUGGCUUUUUUCGGCUGGCUGUAUUUGAAGCAAGCACUUCAAAAGCAGCGUUAGAUUGCAUAAAAGCAAAUGCGGAAGUGGUAA ...((((((((((..............(((((......)))............(((((.(((((((......))))))).)))))....))((((......))))..))))))))))... ( -34.60) >DroYak_CAF1 159178 100 - 1 ACUUUUUUUUUGGUUUU-------UUUUCG-------------GUUUCGACUGGCUGUAUUUGAAGCAAGCACUUCAAAAGCAGCGUUAGAUUGCAUAAAAGCAAAUGCGGAAGUGGUAA ((((((..((((.((((-------....((-------------((((.(((..(((((.(((((((......))))))).))))))))))))))...)))).))))...))))))..... ( -25.60) >consensus AUUUUAUUUCCGCUU____________________________UUUUCGGCUGCCUGUAUUUGAACCAAGCACUUCAAAAGCAGCGUUAGAUUGCAUAAAAGCAAAUGCGGAAGUGGUAA ...((((((((((................................((..((.(.((((.((((((........)))))).)))))))..))((((......))))..))))))))))... (-23.44 = -23.36 + -0.08)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:25:14 2006