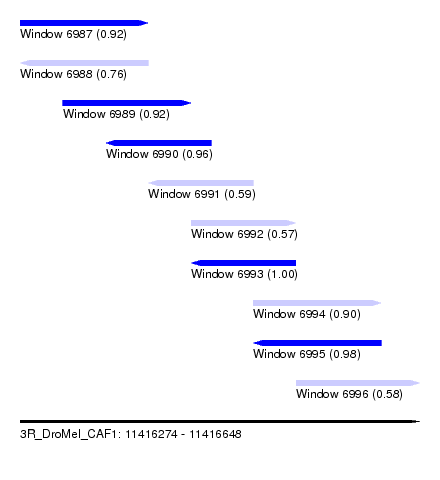
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 11,416,274 – 11,416,648 |
| Length | 374 |
| Max. P | 0.999724 |

| Location | 11,416,274 – 11,416,394 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.57 |
| Mean single sequence MFE | -31.12 |
| Consensus MFE | -22.42 |
| Energy contribution | -23.22 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.75 |
| Structure conservation index | 0.72 |
| SVM decision value | 1.14 |
| SVM RNA-class probability | 0.921021 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11416274 120 + 27905053 CAUCCACAUAUUGUUGGCAUGGCUCUCCAAUUUCUUUUAGUGCACCUCUCUUCGUUUUUGGGGCCAUAGCUCCUUCCACCGAAGUUUGGCAUUUGCCAAACAUUUGCAUAUGGGACCAAC ............(((((..(((....)))...((((...(((((......((((.....(((((....)))))......))))(((((((....)))))))...)))))..))))))))) ( -32.80) >DroSec_CAF1 163904 120 + 1 CAGUCACAUAUUGUUAGAAUGGAUCUCCAAUUUCUUUUAGUGCACCACUCUUCGCUUUUGGGGCCAUAGCUCCUUCCACCAAAGUUUGGCAUUUGCCAAACAUUUGCAUAUGGGACCAAC ..(((.(((((((..(((((((....)))..))))..)))((((...............(((((....)))))..........(((((((....)))))))...)))))))).))).... ( -31.50) >DroSim_CAF1 156699 120 + 1 CAGCCACAUAUUGUUUGAAUGGAUCUCAAAUUUCUUUUAGUGCACCUCUCUUCGUUUUUGGGGCCAUAGCUCCUUCCACCAAAGUUUGGCAUUUGCCAAACAUUUGCAUAUGGGACCAAC ...................(((.(((((((......)).(((((...............(((((....)))))..........(((((((....)))))))...))))).)))))))).. ( -28.50) >DroEre_CAF1 151672 116 + 1 CAGCCACAUACUUUGGUG---GCUCUCCCAUUUCUUUCAGUGCACCUCUCUUCGUUUUC-GGGCCACAGCUCCUUCCACCGAAGUUUGGCAUUUGCCAAACAUUUGCAUAUGGGACCAAC .((((((........)))---))).((((((........(((((............(((-((................)))))(((((((....)))))))...)))))))))))..... ( -35.79) >DroYak_CAF1 155845 119 + 1 CAGCCAAAUAUUCUUGUUAGAUCUCUCCCAUUUCUUUUAGUGCACAUCUCUUCGUUUUC-GGGCCAUAGCUCCUUCCACCGAAGUUUGGCAUUUGCCAAACAUUUGCAUAUGGGACCAAC .........................((((((........(((((............(((-((................)))))(((((((....)))))))...)))))))))))..... ( -26.99) >consensus CAGCCACAUAUUGUUGGAAUGGCUCUCCAAUUUCUUUUAGUGCACCUCUCUUCGUUUUUGGGGCCAUAGCUCCUUCCACCGAAGUUUGGCAUUUGCCAAACAUUUGCAUAUGGGACCAAC ................................((((...(((((......((((.....(((((....)))))......))))(((((((....)))))))...)))))..))))..... (-22.42 = -23.22 + 0.80)

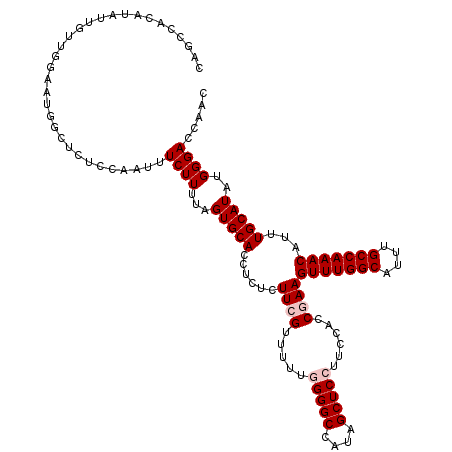
| Location | 11,416,274 – 11,416,394 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.57 |
| Mean single sequence MFE | -34.15 |
| Consensus MFE | -24.92 |
| Energy contribution | -25.16 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.760599 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11416274 120 - 27905053 GUUGGUCCCAUAUGCAAAUGUUUGGCAAAUGCCAAACUUCGGUGGAAGGAGCUAUGGCCCCAAAAACGAAGAGAGGUGCACUAAAAGAAAUUGGAGAGCCAUGCCAACAAUAUGUGGAUG ....(((((((((((....(((((((....)))))))....)).....(.((.((((((((((..........((.....))........)))).).))))))))....))))).)))). ( -32.67) >DroSec_CAF1 163904 120 - 1 GUUGGUCCCAUAUGCAAAUGUUUGGCAAAUGCCAAACUUUGGUGGAAGGAGCUAUGGCCCCAAAAGCGAAGAGUGGUGCACUAAAAGAAAUUGGAGAUCCAUUCUAACAAUAUGUGACUG ...((((.(((((((....(((((((....)))))))(((((.((..(......)..))))))).))..(((((((..(.((((......)))).)..)))))))....))))).)))). ( -36.90) >DroSim_CAF1 156699 120 - 1 GUUGGUCCCAUAUGCAAAUGUUUGGCAAAUGCCAAACUUUGGUGGAAGGAGCUAUGGCCCCAAAAACGAAGAGAGGUGCACUAAAAGAAAUUUGAGAUCCAUUCAAACAAUAUGUGGCUG ...((((.(((((.((((.(((((((....)))))))))))(((((.((.((....)).))...................((.(((....))).)).))))).......))))).)))). ( -31.20) >DroEre_CAF1 151672 116 - 1 GUUGGUCCCAUAUGCAAAUGUUUGGCAAAUGCCAAACUUCGGUGGAAGGAGCUGUGGCCC-GAAAACGAAGAGAGGUGCACUGAAAGAAAUGGGAGAGC---CACCAAAGUAUGUGGCUG .....((((((........(((((((....)))))))(((((((...((.((....))))-....((........)).)))))))....)))))).(((---(((........)))))). ( -39.10) >DroYak_CAF1 155845 119 - 1 GUUGGUCCCAUAUGCAAAUGUUUGGCAAAUGCCAAACUUCGGUGGAAGGAGCUAUGGCCC-GAAAACGAAGAGAUGUGCACUAAAAGAAAUGGGAGAGAUCUAACAAGAAUAUUUGGCUG (((((((((((.((((....((((((....))))))(((((......((.((....))))-.....))))).....))))(.....)..)))))).....)))))............... ( -30.90) >consensus GUUGGUCCCAUAUGCAAAUGUUUGGCAAAUGCCAAACUUCGGUGGAAGGAGCUAUGGCCCCAAAAACGAAGAGAGGUGCACUAAAAGAAAUUGGAGAGCCAUUCCAACAAUAUGUGGCUG ...((((.((((((((....((((((....))))))(((((......((.((....))))......))))).....))).((..((....))..)).............))))).)))). (-24.92 = -25.16 + 0.24)


| Location | 11,416,314 – 11,416,434 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.56 |
| Mean single sequence MFE | -30.67 |
| Consensus MFE | -27.88 |
| Energy contribution | -28.04 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.12 |
| SVM RNA-class probability | 0.917814 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11416314 120 + 27905053 UGCACCUCUCUUCGUUUUUGGGGCCAUAGCUCCUUCCACCGAAGUUUGGCAUUUGCCAAACAUUUGCAUAUGGGACCAACUGUUUUGAAUAUUGAAAGCAGACAACCAACCAACUGCAGG ..........((((.....(((((....)))))......))))(((((((....)))))))..(((((..(((......(((((((........)))))))........)))..))))). ( -32.74) >DroSec_CAF1 163944 120 + 1 UGCACCACUCUUCGCUUUUGGGGCCAUAGCUCCUUCCACCAAAGUUUGGCAUUUGCCAAACAUUUGCAUAUGGGACCAACUGUUUUGAAUAUUGAAAGCAGACAACCAUCGCACUGCCGG (((..........((....(((((....)))))..........(((((((....)))))))....))..((((......(((((((........)))))))....)))).)))....... ( -30.80) >DroSim_CAF1 156739 120 + 1 UGCACCUCUCUUCGUUUUUGGGGCCAUAGCUCCUUCCACCAAAGUUUGGCAUUUGCCAAACAUUUGCAUAUGGGACCAACUGUUUUGAAUAUUGAAAGCAGACAACCAACCAACUGCAUG ((((...............(((((....)))))(((((.(((((((((((....))))))).))))....)))))....(((((((........))))))).............)))).. ( -31.00) >DroEre_CAF1 151709 115 + 1 UGCACCUCUCUUCGUUUUC-GGGCCACAGCUCCUUCCACCGAAGUUUGGCAUUUGCCAAACAUUUGCAUAUGGGACCAACUGUUUUGAAUAUUGAAAGCAGACAACCA----ACUGCGGG ....((....((((..(((-(((..((((.((((.....(((((((((((....))))))).)))).....))))....))))))))))...)))).((((.......----.)))))). ( -30.30) >DroYak_CAF1 155885 115 + 1 UGCACAUCUCUUCGUUUUC-GGGCCAUAGCUCCUUCCACCGAAGUUUGGCAUUUGCCAAACAUUUGCAUAUGGGACCAACUGUUUUGAAUAUUGAAAGCAGACAACCA----ACUGCUGG .(((...............-((.(((((((...(((....)))(((((((....)))))))....)).)))))..))..(((((((........))))))).......----..)))... ( -28.50) >consensus UGCACCUCUCUUCGUUUUUGGGGCCAUAGCUCCUUCCACCGAAGUUUGGCAUUUGCCAAACAUUUGCAUAUGGGACCAACUGUUUUGAAUAUUGAAAGCAGACAACCA_C__ACUGCAGG .(((...............(((((....)))))(((((.(((((((((((....))))))).))))....)))))....(((((((........))))))).............)))... (-27.88 = -28.04 + 0.16)

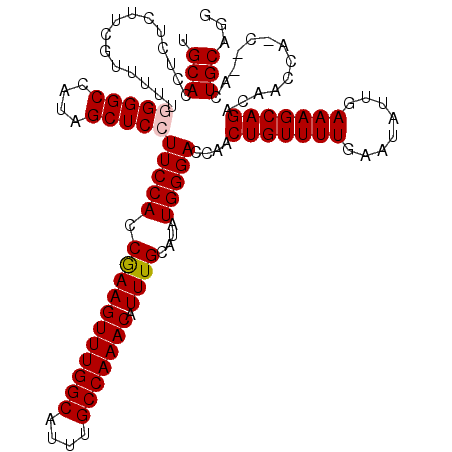
| Location | 11,416,354 – 11,416,453 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.62 |
| Mean single sequence MFE | -26.56 |
| Consensus MFE | -20.20 |
| Energy contribution | -20.04 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.56 |
| SVM RNA-class probability | 0.963708 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11416354 99 - 27905053 ---U----CCCCCAGCUGCCC--------------CAUCCCCUGCAGUUGGUUGGUUGUCUGCUUUCAAUAUUCAAAACAGUUGGUCCCAUAUGCAAAUGUUUGGCAAAUGCCAAACUUC ---.----((.((((((((..--------------........))))))))..)).....(((.........((((.....))))........)))...(((((((....)))))))... ( -24.63) >DroSec_CAF1 163984 115 - 1 CAAC----CCCCCAGCUGCC-CACAACCCCCCGCUCAACCCCGGCAGUGCGAUGGUUGUCUGCUUUCAAUAUUCAAAACAGUUGGUCCCAUAUGCAAAUGUUUGGCAAAUGCCAAACUUU ....----...(((((((..-.((((((...(((.(..(....)..).)))..)))))).((....))..........)))))))..............(((((((....)))))))... ( -28.10) >DroSim_CAF1 156779 116 - 1 CAAC----CCCCUAGCUGCCCCUCAACCCCCCGCUCAUCCCAUGCAGUUGGUUGGUUGUCUGCUUUCAAUAUUCAAAACAGUUGGUCCCAUAUGCAAAUGUUUGGCAAAUGCCAAACUUU ((((----(..((((((((........................))))))))..)))))..(((.........((((.....))))........)))...(((((((....)))))))... ( -26.99) >DroEre_CAF1 151748 115 - 1 CCCCAGCACCCCCAGCUGCCC-CAAGCCCCCUGCUCCACCCCCGCAGU----UGGUUGUCUGCUUUCAAUAUUCAAAACAGUUGGUCCCAUAUGCAAAUGUUUGGCAAAUGCCAAACUUC ..(((((....((((((((..-..(((.....)))........)))))----)))(((........)))...........)))))..............(((((((....)))))))... ( -28.40) >DroYak_CAF1 155924 106 - 1 ---------CCCCAGCUGCCC-CAAACCCCCCGCUCCACCCCAGCAGU----UGGUUGUCUGCUUUCAAUAUUCAAAACAGUUGGUCCCAUAUGCAAAUGUUUGGCAAAUGCCAAACUUC ---------..(((((((...-..........(((.......))).((----(((..(....)..)))))........)))))))..............(((((((....)))))))... ( -24.70) >consensus C__C____CCCCCAGCUGCCC_CAAACCCCCCGCUCAACCCCUGCAGU__G_UGGUUGUCUGCUUUCAAUAUUCAAAACAGUUGGUCCCAUAUGCAAAUGUUUGGCAAAUGCCAAACUUC ...........(((((((.........................((((............))))(((........))).)))))))..............(((((((....)))))))... (-20.20 = -20.04 + -0.16)

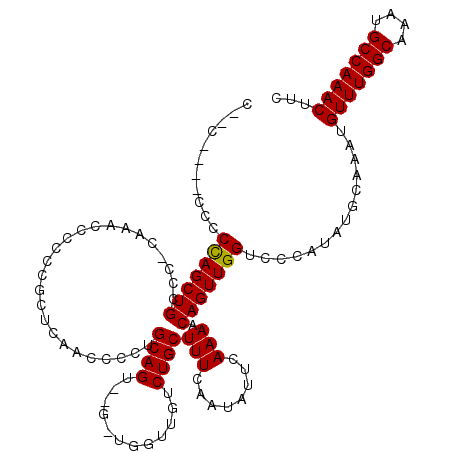

| Location | 11,416,394 – 11,416,492 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.09 |
| Mean single sequence MFE | -22.21 |
| Consensus MFE | -13.97 |
| Energy contribution | -13.97 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.586233 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11416394 98 - 27905053 GCAACUCUGUUAACCCUGCCACACGCAAUCACCGCAGCA----U----CCCCCAGCUGCCC--------------CAUCCCCUGCAGUUGGUUGGUUGUCUGCUUUCAAUAUUCAAAACA ........(((.....(((.....)))......((((((----.----((.((((((((..--------------........))))))))..)).)).)))).............))). ( -23.20) >DroSec_CAF1 164024 115 - 1 GCAACUCUGUUAACCCUGCCACACGCAAUCACCGCAGCAGCAAC----CCCCCAGCUGCC-CACAACCCCCCGCUCAACCCCGGCAGUGCGAUGGUUGUCUGCUUUCAAUAUUCAAAACA ........(((.....(((.....)))........(((((((((----(.....((((((-.....................)))))).....))))).)))))............))). ( -24.50) >DroSim_CAF1 156819 116 - 1 GCAACUCUGUUAACCCUGCCACACGCAAUCACCGCAGCAGCAAC----CCCCUAGCUGCCCCUCAACCCCCCGCUCAUCCCAUGCAGUUGGUUGGUUGUCUGCUUUCAAUAUUCAAAACA ........(((.....(((.....)))........(((((((((----(..((((((((........................))))))))..))))).)))))............))). ( -25.06) >DroEre_CAF1 151788 115 - 1 GCAACUCUGUUAACCCUGCCACACGCAAUCACCGCAGCACCCCCAGCACCCCCAGCUGCCC-CAAGCCCCCUGCUCCACCCCCGCAGU----UGGUUGUCUGCUUUCAAUAUUCAAAACA ........(((.....(((.....)))......((((.((...((((.......))))...-..((((..((((.........)))).----.)))))))))).............))). ( -19.70) >DroYak_CAF1 155964 104 - 1 GCAACUCUGUUAACCCUGCCACACGCAAUCACCGCAGC-----------CCCCAGCUGCCC-CAAACCCCCCGCUCCACCCCAGCAGU----UGGUUGUCUGCUUUCAAUAUUCAAAACA ........(((.....(((.....)))......(((((-----------..((((((((..-.....................)))))----)))..).)))).............))). ( -18.60) >consensus GCAACUCUGUUAACCCUGCCACACGCAAUCACCGCAGCA_C__C____CCCCCAGCUGCCC_CAAACCCCCCGCUCAACCCCUGCAGU__G_UGGUUGUCUGCUUUCAAUAUUCAAAACA ((((((((((......(((.....)))......(((((................)))))........................))))......))))))..................... (-13.97 = -13.97 + -0.00)

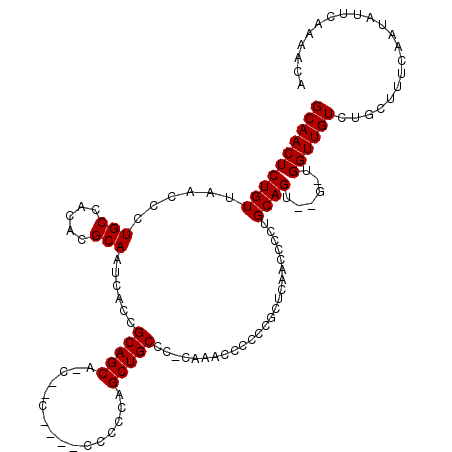
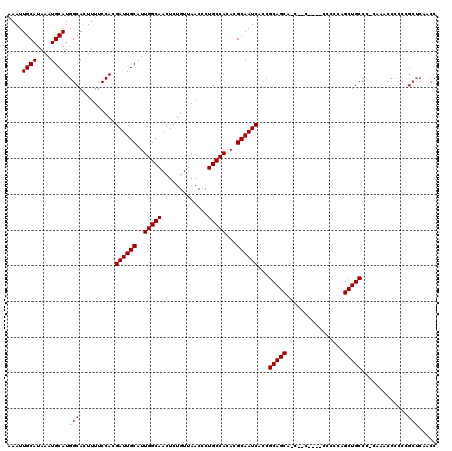
| Location | 11,416,434 – 11,416,532 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.83 |
| Mean single sequence MFE | -36.18 |
| Consensus MFE | -29.50 |
| Energy contribution | -29.70 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.567008 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11416434 98 + 27905053 GGAUG--------------GGGCAGCUGGGGG----A----UGCUGCGGUGAUUGCGUGUGGCAGGGUUAACAGAGUUGCCAAUGCAAUCGUGGAAAAGUGCCAUGCAUUUAUGCAAUUU (.(((--------------(.((.((((.((.----.----..)).))))((((((((.((((((..(.....)..))))))))))))))........)).)))).)............. ( -32.60) >DroSec_CAF1 164064 115 + 1 GGUUGAGCGGGGGGUUGUG-GGCAGCUGGGGG----GUUGCUGCUGCGGUGAUUGCGUGUGGCAGGGUUAACAGAGUUGCCAAUGCAAUCGUGGAAAAGUGCCAUGCAUUUAUGCAAUUU ......(((...((((((.-(((((((..(..----(.(.((((..((((....)).))..)))).).)..)..)))))))...))))))......((((((...)))))).)))..... ( -36.80) >DroSim_CAF1 156859 116 + 1 GGAUGAGCGGGGGGUUGAGGGGCAGCUAGGGG----GUUGCUGCUGCGGUGAUUGCGUGUGGCAGGGUUAACAGAGUUGCCAAUGCAAUCGUGGAAAAGUGCCAUGCAUUUAUGCAAUUU (.(((((((((.(.((.((.(((((((....)----)))))).)).(.((((((((((.((((((..(.....)..)))))))))))))))).)..)).).)).)))..)))).)..... ( -36.80) >DroEre_CAF1 151824 119 + 1 GGUGGAGCAGGGGGCUUG-GGGCAGCUGGGGGUGCUGGGGGUGCUGCGGUGAUUGCGUGUGGCAGGGUUAACAGAGUUGCCAAUGCAAUCGUGGAAAAGUGCCAUGCAUUUAUGCAAUUU ..(.((((.....)))).-)....((...((((((....((..((.(.((((((((((.((((((..(.....)..)))))))))))))))).)...))..))..))))))..))..... ( -40.00) >DroYak_CAF1 156000 108 + 1 GGUGGAGCGGGGGGUUUG-GGGCAGCUGGGG-----------GCUGCGGUGAUUGCGUGUGGCAGGGUUAACAGAGUUGCCAAUGCAAUCGUGGAAAAGUGCCAUGCAUUUAUGCAAUUU .((((((((((.(.(((.-..(((((.....-----------)))))...((((((((.((((((..(.....)..)))))))))))))).....))).).)).))).)))))....... ( -34.70) >consensus GGUUGAGCGGGGGGUUGG_GGGCAGCUGGGGG____G__G_UGCUGCGGUGAUUGCGUGUGGCAGGGUUAACAGAGUUGCCAAUGCAAUCGUGGAAAAGUGCCAUGCAUUUAUGCAAUUU ....................((((((((.((............)).))))((((((((.((((((..(.....)..)))))))))))))).........)))).((((....)))).... (-29.50 = -29.70 + 0.20)


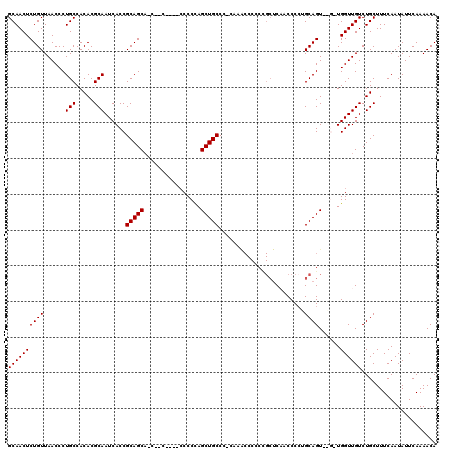
| Location | 11,416,434 – 11,416,532 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.83 |
| Mean single sequence MFE | -26.54 |
| Consensus MFE | -22.79 |
| Energy contribution | -23.39 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.59 |
| Structure conservation index | 0.86 |
| SVM decision value | 3.95 |
| SVM RNA-class probability | 0.999724 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11416434 98 - 27905053 AAAUUGCAUAAAUGCAUGGCACUUUUCCACGAUUGCAUUGGCAACUCUGUUAACCCUGCCACACGCAAUCACCGCAGCA----U----CCCCCAGCUGCCC--------------CAUCC ....((((....))))(((...........((((((..(((((.............)))))...))))))...(((((.----.----......))))).)--------------))... ( -26.02) >DroSec_CAF1 164064 115 - 1 AAAUUGCAUAAAUGCAUGGCACUUUUCCACGAUUGCAUUGGCAACUCUGUUAACCCUGCCACACGCAAUCACCGCAGCAGCAAC----CCCCCAGCUGCC-CACAACCCCCCGCUCAACC .....((.....(((.(((.......))).((((((..(((((.............)))))...))))))...)))(((((...----......))))).-...........))...... ( -26.42) >DroSim_CAF1 156859 116 - 1 AAAUUGCAUAAAUGCAUGGCACUUUUCCACGAUUGCAUUGGCAACUCUGUUAACCCUGCCACACGCAAUCACCGCAGCAGCAAC----CCCCUAGCUGCCCCUCAACCCCCCGCUCAUCC .....((.....(((.(((.......))).((((((..(((((.............)))))...))))))...)))(((((...----......))))).............))...... ( -26.42) >DroEre_CAF1 151824 119 - 1 AAAUUGCAUAAAUGCAUGGCACUUUUCCACGAUUGCAUUGGCAACUCUGUUAACCCUGCCACACGCAAUCACCGCAGCACCCCCAGCACCCCCAGCUGCCC-CAAGCCCCCUGCUCCACC ....((((....)))).(((..........((((((..(((((.............)))))...))))))...(((((................)))))..-...)))............ ( -27.51) >DroYak_CAF1 156000 108 - 1 AAAUUGCAUAAAUGCAUGGCACUUUUCCACGAUUGCAUUGGCAACUCUGUUAACCCUGCCACACGCAAUCACCGCAGC-----------CCCCAGCUGCCC-CAAACCCCCCGCUCCACC ....((((....)))).(((..........((((((..(((((.............)))))...))))))...(((((-----------.....)))))..-..........)))..... ( -26.32) >consensus AAAUUGCAUAAAUGCAUGGCACUUUUCCACGAUUGCAUUGGCAACUCUGUUAACCCUGCCACACGCAAUCACCGCAGCA_C__C____CCCCCAGCUGCCC_CAAACCCCCCGCUCAACC ....((((....)))).(((..........((((((..(((((.............)))))...))))))...(((((................))))).............)))..... (-22.79 = -23.39 + 0.60)

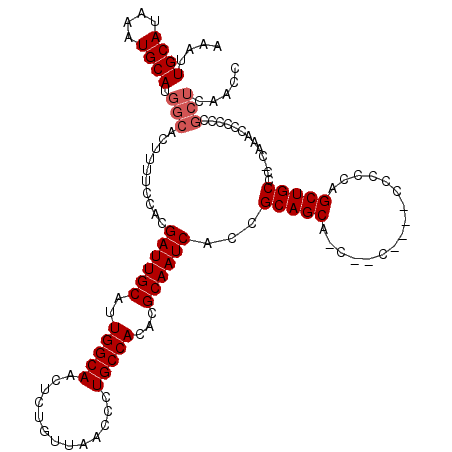
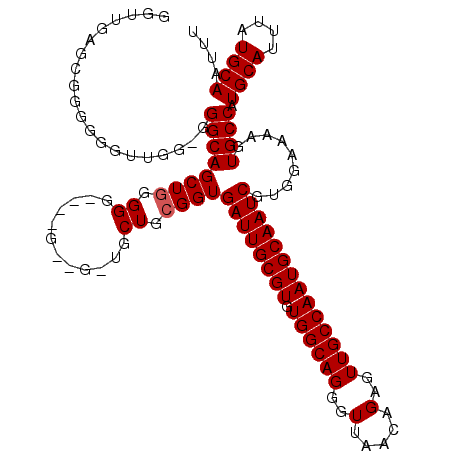
| Location | 11,416,492 – 11,416,612 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.12 |
| Mean single sequence MFE | -33.66 |
| Consensus MFE | -30.96 |
| Energy contribution | -31.44 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.37 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.897600 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11416492 120 + 27905053 CAAUGCAAUCGUGGAAAAGUGCCAUGCAUUUAUGCAAUUUUCACUGACUUGGCUUGCUGCUCGAAAGCCACUCAAAUGCCAUGGCAGGGCACUCCAUCAAGUGCAGUUACCUCGCAGCCC ...((((...(((((...(((((.((((....))))........(((..((((((.........)))))).)))..(((....))).))))))))))....))))............... ( -32.70) >DroSec_CAF1 164139 119 + 1 CAAUGCAAUCGUGGAAAAGUGCCAUGCAUUUAUGCAAUUUUCACUGACUUGGCUUGCUGCUCGAAAGCCACUCAAAUGCCAUGGCAGGGCACUCCAUCAAGUGCAGUUGCCUCGCAGCC- ...((((...(((((...(((((.((((....))))........(((..((((((.........)))))).)))..(((....))).))))))))))....))))(((((...))))).- ( -36.60) >DroSim_CAF1 156935 119 + 1 CAAUGCAAUCGUGGAAAAGUGCCAUGCAUUUAUGCAAUUUUCACUGACUUGGCUUGCUGCUCGAAAGCCACUCAAAUGCCAUGGCAGGGCACUCCAUCAAGUGCAGUUGCCUCGCAGCC- ...((((...(((((...(((((.((((....))))........(((..((((((.........)))))).)))..(((....))).))))))))))....))))(((((...))))).- ( -36.60) >DroEre_CAF1 151903 114 + 1 CAAUGCAAUCGUGGAAAAGUGCCAUGCAUUUAUGCAAUUUUCACCGACUUGGCUUGCUGCUCGAAAGCCACUCAAAUGCCAUGGCAGGGCACUCCAUCAAGUGCAGUUG-----CAACC- (((((((...(((((...(((((.((((....)))).........((..((((((.........)))))).))...(((....))).))))))))))....)))).)))-----.....- ( -32.30) >DroYak_CAF1 156068 114 + 1 CAAUGCAAUCGUGGAAAAGUGCCAUGCAUUUAUGCAAUUUUCACCGACUUGGCUUGCUGCUCGAAAGCCACUCAAAUGCCAUGGCAGAGCACUCCAUCAAGUGCAGCUG-----CAACC- ...((((............((((((((((((.((.......))..((..((((((.........)))))).))))))).)))))))..(((((......)))))...))-----))...- ( -30.10) >consensus CAAUGCAAUCGUGGAAAAGUGCCAUGCAUUUAUGCAAUUUUCACUGACUUGGCUUGCUGCUCGAAAGCCACUCAAAUGCCAUGGCAGGGCACUCCAUCAAGUGCAGUUGCCUCGCAGCC_ ...((((...(((((...(((((.((((....)))).........((..((((((.........)))))).))...(((....))).))))))))))....))))((((.....)))).. (-30.96 = -31.44 + 0.48)

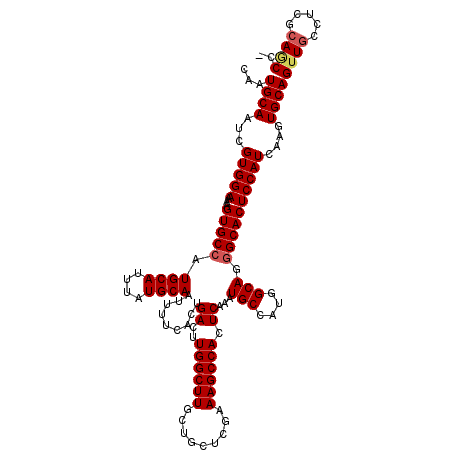
| Location | 11,416,492 – 11,416,612 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.12 |
| Mean single sequence MFE | -38.98 |
| Consensus MFE | -38.14 |
| Energy contribution | -37.58 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.98 |
| SVM decision value | 1.88 |
| SVM RNA-class probability | 0.981075 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11416492 120 - 27905053 GGGCUGCGAGGUAACUGCACUUGAUGGAGUGCCCUGCCAUGGCAUUUGAGUGGCUUUCGAGCAGCAAGCCAAGUCAGUGAAAAUUGCAUAAAUGCAUGGCACUUUUCCACGAUUGCAUUG ..((..((.((.....((((((....))))))..(((((((.((((((..((((((.(.....).)))))).(.((((....))))).))))))))))))).....)).))...)).... ( -39.10) >DroSec_CAF1 164139 119 - 1 -GGCUGCGAGGCAACUGCACUUGAUGGAGUGCCCUGCCAUGGCAUUUGAGUGGCUUUCGAGCAGCAAGCCAAGUCAGUGAAAAUUGCAUAAAUGCAUGGCACUUUUCCACGAUUGCAUUG -.(((....)))...((((.(((.(((((.....(((((((.((((((..((((((.(.....).)))))).(.((((....))))).)))))))))))))...)))))))).))))... ( -40.40) >DroSim_CAF1 156935 119 - 1 -GGCUGCGAGGCAACUGCACUUGAUGGAGUGCCCUGCCAUGGCAUUUGAGUGGCUUUCGAGCAGCAAGCCAAGUCAGUGAAAAUUGCAUAAAUGCAUGGCACUUUUCCACGAUUGCAUUG -.(((....)))...((((.(((.(((((.....(((((((.((((((..((((((.(.....).)))))).(.((((....))))).)))))))))))))...)))))))).))))... ( -40.40) >DroEre_CAF1 151903 114 - 1 -GGUUG-----CAACUGCACUUGAUGGAGUGCCCUGCCAUGGCAUUUGAGUGGCUUUCGAGCAGCAAGCCAAGUCGGUGAAAAUUGCAUAAAUGCAUGGCACUUUUCCACGAUUGCAUUG -.....-----(((.((((.(((.(((((.....(((((((.((((((..((((((.(.....).)))))).(.((((....))))).)))))))))))))...)))))))).))))))) ( -37.50) >DroYak_CAF1 156068 114 - 1 -GGUUG-----CAGCUGCACUUGAUGGAGUGCUCUGCCAUGGCAUUUGAGUGGCUUUCGAGCAGCAAGCCAAGUCGGUGAAAAUUGCAUAAAUGCAUGGCACUUUUCCACGAUUGCAUUG -.....-----(((.((((.(((.(((((.....(((((((.((((((..((((((.(.....).)))))).(.((((....))))).)))))))))))))...)))))))).))))))) ( -37.50) >consensus _GGCUGCGAGGCAACUGCACUUGAUGGAGUGCCCUGCCAUGGCAUUUGAGUGGCUUUCGAGCAGCAAGCCAAGUCAGUGAAAAUUGCAUAAAUGCAUGGCACUUUUCCACGAUUGCAUUG ...........(((.((((.(((.(((((.....(((((((.((((((..((((((.(.....).)))))).(.((((....))))).)))))))))))))...)))))))).))))))) (-38.14 = -37.58 + -0.56)

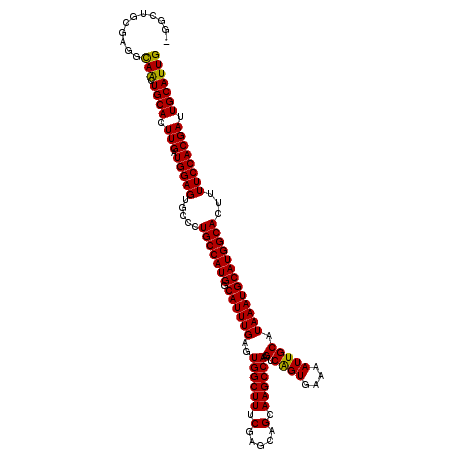
| Location | 11,416,532 – 11,416,648 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.34 |
| Mean single sequence MFE | -32.32 |
| Consensus MFE | -18.76 |
| Energy contribution | -19.64 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.578350 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11416532 116 + 27905053 UCACUGACUUGGCUUGCUGCUCGAAAGCCACUCAAAUGCCAUGGCAGGGCACUCCAUCAAGUGCAGUUACCUCGCAGCCCCC-UCCAGCAACCAACUUGCAACCCC---CUUUGGCCGUG .((((((..((((((.........)))))).)))...(((((((.((((((((......))))).((......)).....))-))))((((.....))))......---...)))).))) ( -29.90) >DroSec_CAF1 164179 115 + 1 UCACUGACUUGGCUUGCUGCUCGAAAGCCACUCAAAUGCCAUGGCAGGGCACUCCAUCAAGUGCAGUUGCCUCGCAGCC-CC-UCCAGCAAGCAACUUGCAACCCC---CUUUUGCAGUG .((((......((((((((..(....).........(((...(((((.(((((......)))))..)))))..)))...-..-..)))))))).....((((....---...)))))))) ( -35.70) >DroSim_CAF1 156975 115 + 1 UCACUGACUUGGCUUGCUGCUCGAAAGCCACUCAAAUGCCAUGGCAGGGCACUCCAUCAAGUGCAGUUGCCUCGCAGCC-CC-UCCAGCAAGCAACUUGCAACCCC---CUUCGUCAGUG .(((((((.(((...(((((.(....)...............(((((.(((((......)))))..)))))..))))).-..-.)))(((((...)))))......---....))))))) ( -39.60) >DroEre_CAF1 151943 95 + 1 UCACCGACUUGGCUUGCUGCUCGAAAGCCACUCAAAUGCCAUGGCAGGGCACUCCAUCAAGUGCAGUUG-----CAACC-CC--U----------CGUGCAACACCACCAUGC------- .....((..((((((.........)))))).))......(((((..(((((((......))))).((((-----((...-..--.----------..)))))).)).))))).------- ( -28.80) >DroYak_CAF1 156108 98 + 1 UCACCGACUUGGCUUGCUGCUCGAAAGCCACUCAAAUGCCAUGGCAGAGCACUCCAUCAAGUGCAGCUG-----CAACC-CCCUU----------CGUGCAACCCC---CUGCGGCA--- .....((..((((((.........)))))).))...((((...((((.(((((......)))))..)))-----)....-.....----------...(((.....---.)))))))--- ( -27.60) >consensus UCACUGACUUGGCUUGCUGCUCGAAAGCCACUCAAAUGCCAUGGCAGGGCACUCCAUCAAGUGCAGUUGCCUCGCAGCC_CC_UCCAGCAA_CAACUUGCAACCCC___CUUCGGCAGUG .....((..((((((.........)))))).))..........((((((((((......))))).((((.....)))).................))))).................... (-18.76 = -19.64 + 0.88)

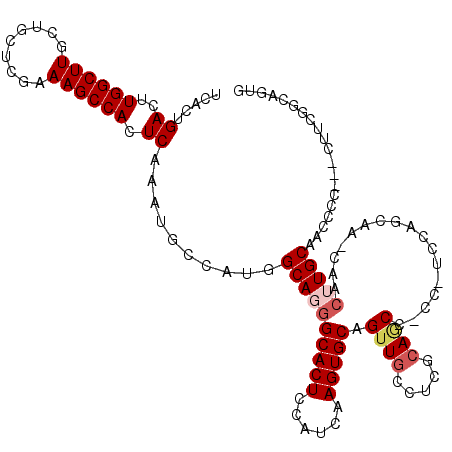
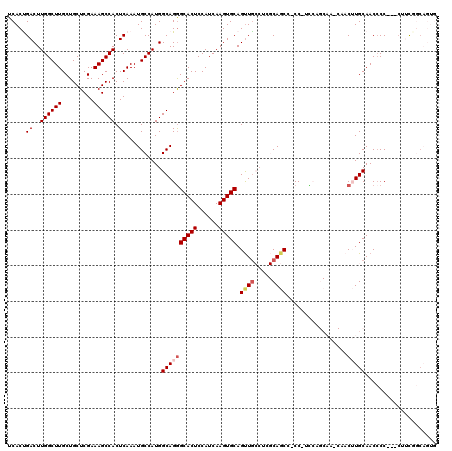
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:25:06 2006