
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 11,415,892 – 11,416,080 |
| Length | 188 |
| Max. P | 0.996066 |

| Location | 11,415,892 – 11,416,011 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.49 |
| Mean single sequence MFE | -42.10 |
| Consensus MFE | -39.92 |
| Energy contribution | -40.36 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.22 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.688421 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11415892 119 + 27905053 CCACCCAGUCAUUACCAUCAAUUUCCAGCCCCAAGGGCAGUGGACAA-GUUCGGGUUUUGGCACAUGGGUCAUUUGUGUGCAUUUGUUUUCAGGGUGUCCCCGGCAGGAUCCUGCUGGCC .(((((...((..(((...((((((((((((...))))..))))..)-)))..)))..))(((((..(.....)..)))))...........)))))...(((((((....))))))).. ( -44.10) >DroSec_CAF1 163534 119 + 1 CCACCCAGUCAUUACCAUCAAUUUCCAGCCCCAAGGGCAGUGGACAA-GUUCGGGUUUUGGCACAUGGGUCAUUUGUGUGCAUUUGUUUUCAGGGUGUCCCCGGCAGGAUCCUGCUGGCC .(((((...((..(((...((((((((((((...))))..))))..)-)))..)))..))(((((..(.....)..)))))...........)))))...(((((((....))))))).. ( -44.10) >DroSim_CAF1 156320 119 + 1 CCACCCAGUCAUUACCAUCAAUUUCCAGCCCCAAGGGCAGUGGACAA-GUUCGGGUUUUGGCACAUGGGUCAUUUGUGUGCAUUUGUUUUCAGGGUUUCCCCGGCAGGAUCCUGCUGGCC ..((((...((..(((...((((((((((((...))))..))))..)-)))..)))..))(((((..(.....)..)))))...........))))....(((((((....))))))).. ( -41.00) >DroEre_CAF1 151324 120 + 1 CCAGCCAGUCAUUACCAUCAAUUUGCAGCCCCAAGGGCAGCGGACCAGGUUCGGGUUUUGGCACAUGGGUCAUUUGUGUGCAUUUGUUUUCAGGGUGUCCCCGGCAGGAUCCUGCUGGCC ...((((((....(((.....(((((.((((...)))).)))))...)))..(((((((((((((..(.....)..)))))...........(((....)))..)))))))).)))))). ( -42.10) >DroYak_CAF1 155501 120 + 1 CCACCCAGUCAUUACCAUCAAUUUCCAGCCCCAAGGGCAGUGGACAAGGUUCGGGUUUUGGCACAUGGGUCAUUUGUGUGCAUUUGUUUUCAGGGUGUCCCCGGCUGGAUCCUGCUGGCC .(((((...((..(((..(....((((((((...))))..))))....)....)))..))(((((..(.....)..)))))...........)))))...(((((.(....).))))).. ( -39.20) >consensus CCACCCAGUCAUUACCAUCAAUUUCCAGCCCCAAGGGCAGUGGACAA_GUUCGGGUUUUGGCACAUGGGUCAUUUGUGUGCAUUUGUUUUCAGGGUGUCCCCGGCAGGAUCCUGCUGGCC .(((((...((..(((...........((((...))))..(((((...))))))))..))(((((..(.....)..)))))...........)))))...(((((((....))))))).. (-39.92 = -40.36 + 0.44)

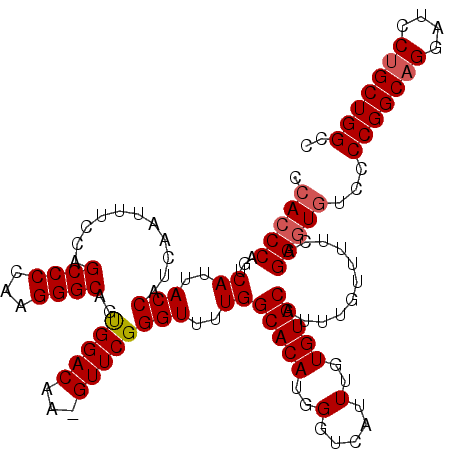

| Location | 11,415,932 – 11,416,046 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.40 |
| Mean single sequence MFE | -35.37 |
| Consensus MFE | -32.30 |
| Energy contribution | -32.30 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.91 |
| SVM decision value | 2.65 |
| SVM RNA-class probability | 0.996066 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11415932 114 - 27905053 AAUCAAC-----GGCUGCCGGGCAGGAUAAGCAAUGUCCUGGCCAGCAGGAUCCUGCCGGGGACACCCUGAAAACAAAUGCACACAAAUGACCCAUGUGCCAAAACCCGAAC-UUGUCCA ......(-----(((.((.((.(((((((.....))))))).)).))((....)))))).(((((..............(((((...........)))))............-.))))). ( -34.73) >DroSec_CAF1 163574 114 - 1 AAUCAAC-----GGCUGCCGGGCAGGAUAAGCAAUGUCCUGGCCAGCAGGAUCCUGCCGGGGACACCCUGAAAACAAAUGCACACAAAUGACCCAUGUGCCAAAACCCGAAC-UUGUCCA ......(-----(((.((.((.(((((((.....))))))).)).))((....)))))).(((((..............(((((...........)))))............-.))))). ( -34.73) >DroSim_CAF1 156360 114 - 1 AAUCAAC-----GGCUGCCGGGCAGGAUAAGCAAUGUCCUGGCCAGCAGGAUCCUGCCGGGGAAACCCUGAAAACAAAUGCACACAAAUGACCCAUGUGCCAAAACCCGAAC-UUGUCCA ......(-----((((((.((.(((((((.....))))))).)).)))).........(((....)))...........(((((...........)))))......)))...-....... ( -36.10) >DroEre_CAF1 151364 120 - 1 AAUCAACGCAUCGGCUGCCGGGCAGGAUAAGCAAUGUCCUGGCCAGCAGGAUCCUGCCGGGGACACCCUGAAAACAAAUGCACACAAAUGACCCAUGUGCCAAAACCCGAACCUGGUCCG ..........((((((((.((.(((((((.....))))))).)).))))........(((((...))))).........(((((...........)))))......)))).......... ( -35.40) >DroYak_CAF1 155541 115 - 1 AAUCAAC-----GGCUGCCGGGCAGGAUAAGCAAUGUCCUGGCCAGCAGGAUCCAGCCGGGGACACCCUGAAAACAAAUGCACACAAAUGACCCAUGUGCCAAAACCCGAACCUUGUCCA ......(-----((((((.((.(((((((.....))))))).)).)).(....)))))).(((((..............(((((...........)))))..............))))). ( -35.89) >consensus AAUCAAC_____GGCUGCCGGGCAGGAUAAGCAAUGUCCUGGCCAGCAGGAUCCUGCCGGGGACACCCUGAAAACAAAUGCACACAAAUGACCCAUGUGCCAAAACCCGAAC_UUGUCCA ..(((.......((((((.((.(((((((.....))))))).)).)))(....).)))(((....))))))........(((((...........))))).................... (-32.30 = -32.30 + 0.00)

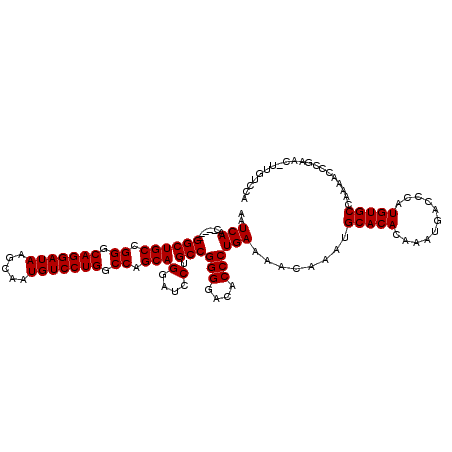

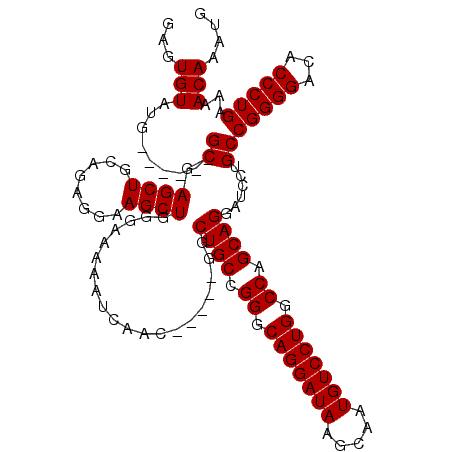
| Location | 11,415,971 – 11,416,080 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.47 |
| Mean single sequence MFE | -34.62 |
| Consensus MFE | -29.70 |
| Energy contribution | -29.58 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.46 |
| SVM RNA-class probability | 0.955897 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11415971 109 + 27905053 CAUUUGUUUUCAGGGUGUCCCCGGCAGGAUCCUGCUGGCCAGGACAUUGCUUAUCCUGCCCGGCAGCC-----GUUGAUUUUUCCCAGCUUCCUCCGCAGCUCGC------AAUACACUC ............((((((....((.((((..(((((((.(((((.(.....).))))).)))))))..-----((((........)))).))))))((.....))------...)))))) ( -35.20) >DroSec_CAF1 163613 109 + 1 CAUUUGUUUUCAGGGUGUCCCCGGCAGGAUCCUGCUGGCCAGGACAUUGCUUAUCCUGCCCGGCAGCC-----GUUGAUUUUUCCCAGCUUCCUCUGCAGCUCGC------CAUACACUC .............((((....(((.((((..(((((((.(((((.(.....).))))).)))))))..-----((((........)))).))))))).....)))------)........ ( -33.30) >DroSim_CAF1 156399 109 + 1 CAUUUGUUUUCAGGGUUUCCCCGGCAGGAUCCUGCUGGCCAGGACAUUGCUUAUCCUGCCCGGCAGCC-----GUUGAUUUUUCCCAGCUUCCUCUGCAGCUUGC------CAUACACUC ............(((....)))((((((...(((((((.(((((.(.....).))))).)))))))..-----((((........))))...........)))))------)........ ( -35.10) >DroEre_CAF1 151404 119 + 1 CAUUUGUUUUCAGGGUGUCCCCGGCAGGAUCCUGCUGGCCAGGACAUUGCUUAUCCUGCCCGGCAGCCGAUGCGUUGAUUUUUC-CAGCUUCCUCUGCAGCUCGCAGUACUCGUACACUC ............(((((((((((((((....)))))))...))))..........((((...((((..((.((...((....))-..)).))..)))).....)))).))))........ ( -35.50) >DroYak_CAF1 155581 108 + 1 CAUUUGUUUUCAGGGUGUCCCCGGCUGGAUCCUGCUGGCCAGGACAUUGCUUAUCCUGCCCGGCAGCC-----GUUGAUUUUUC-CAGCUUCCUCUGCAGCUCGC------CAUACACUC ............((((((...((((((....(((((((.(((((.(.....).))))).)))))))..-----((((.......-))))........)))).)).------...)))))) ( -34.00) >consensus CAUUUGUUUUCAGGGUGUCCCCGGCAGGAUCCUGCUGGCCAGGACAUUGCUUAUCCUGCCCGGCAGCC_____GUUGAUUUUUCCCAGCUUCCUCUGCAGCUCGC______CAUACACUC ............((((.....(((.((((..(((((((.(((((.(.....).))))).))))))).......((((........)))).)))))))..))))................. (-29.70 = -29.58 + -0.12)



| Location | 11,415,971 – 11,416,080 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.47 |
| Mean single sequence MFE | -39.90 |
| Consensus MFE | -32.92 |
| Energy contribution | -32.92 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.83 |
| SVM decision value | 2.05 |
| SVM RNA-class probability | 0.986795 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11415971 109 - 27905053 GAGUGUAUU------GCGAGCUGCGGAGGAAGCUGGGAAAAAUCAAC-----GGCUGCCGGGCAGGAUAAGCAAUGUCCUGGCCAGCAGGAUCCUGCCGGGGACACCCUGAAAACAAAUG ...(((...------((((.((((...((.(((((...........)-----)))).))((.(((((((.....))))))).)).))))..))..))(((((...)))))...))).... ( -36.90) >DroSec_CAF1 163613 109 - 1 GAGUGUAUG------GCGAGCUGCAGAGGAAGCUGGGAAAAAUCAAC-----GGCUGCCGGGCAGGAUAAGCAAUGUCCUGGCCAGCAGGAUCCUGCCGGGGACACCCUGAAAACAAAUG ...(((..(------((((.((((...((.(((((...........)-----)))).))((.(((((((.....))))))).)).))))..))..)))(((....))).....))).... ( -40.20) >DroSim_CAF1 156399 109 - 1 GAGUGUAUG------GCAAGCUGCAGAGGAAGCUGGGAAAAAUCAAC-----GGCUGCCGGGCAGGAUAAGCAAUGUCCUGGCCAGCAGGAUCCUGCCGGGGAAACCCUGAAAACAAAUG ...(((..(------(((.(((((...((.(((((...........)-----)))).))((.(((((((.....))))))).)).))))...).))))(((....))).....))).... ( -43.00) >DroEre_CAF1 151404 119 - 1 GAGUGUACGAGUACUGCGAGCUGCAGAGGAAGCUG-GAAAAAUCAACGCAUCGGCUGCCGGGCAGGAUAAGCAAUGUCCUGGCCAGCAGGAUCCUGCCGGGGACACCCUGAAAACAAAUG ....(((.((...((((.....((((..((.((..-((....))...)).))..)))).((.(((((((.....))))))).)).))))..)).)))(((((...))))).......... ( -39.10) >DroYak_CAF1 155581 108 - 1 GAGUGUAUG------GCGAGCUGCAGAGGAAGCUG-GAAAAAUCAAC-----GGCUGCCGGGCAGGAUAAGCAAUGUCCUGGCCAGCAGGAUCCAGCCGGGGACACCCUGAAAACAAAUG ...(((..(------((((.((((...((.(((((-..........)-----)))).))((.(((((((.....))))))).)).))))..))..)))(((....))).....))).... ( -40.30) >consensus GAGUGUAUG______GCGAGCUGCAGAGGAAGCUGGGAAAAAUCAAC_____GGCUGCCGGGCAGGAUAAGCAAUGUCCUGGCCAGCAGGAUCCUGCCGGGGACACCCUGAAAACAAAUG ...(((.........((.((((........))))....................((((.((.(((((((.....))))))).)).))))......))(((((...)))))...))).... (-32.92 = -32.92 + 0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:24:57 2006