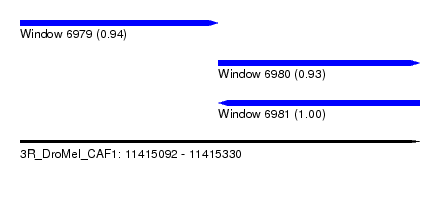
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 11,415,092 – 11,415,330 |
| Length | 238 |
| Max. P | 0.999592 |

| Location | 11,415,092 – 11,415,210 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.32 |
| Mean single sequence MFE | -42.36 |
| Consensus MFE | -31.40 |
| Energy contribution | -33.56 |
| Covariance contribution | 2.16 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.74 |
| SVM decision value | 1.31 |
| SVM RNA-class probability | 0.940656 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11415092 118 + 27905053 CUGUCU--UUUUUUGGGUUUAAAGUUCCGGCAGGUGGGCCAUCGGUCGGUGGACUACCUUCACUUUUUUCGGUGGCUCCUAAUUUUGGGUAGAACCCGAUCUGCACCGGAGCAGCUGCCC ......--......((((.....((((((((((((((((((((((..((((((.....))))))....)))))))))).......((((.....))))))))))..))))))....)))) ( -46.00) >DroSec_CAF1 162672 120 + 1 CUGCCUUUUUUUUUGGGUUUAAAGUUCCGGCAGGUGGGCCAUCGGUCGGUGGACUACCUUCACUUUUUUCGGUGGCUCCUAAUUUUGGGUAGAACCCGAUCUGCACCGGAGCAGCUGCCC ..............((((.....((((((((((((((((((((((..((((((.....))))))....)))))))))).......((((.....))))))))))..))))))....)))) ( -46.00) >DroSim_CAF1 155458 120 + 1 NNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNAGGUGGGCCAUCGGACGGUGGACUACCUUCACUUUUUUCCGUGGCUCCUAAUUUUGGGUAGAACCCGAUCUGCACCGGAGCAGCUGCCC ................................((.((((((.((((.((((((.....))))))....))))))))))))......((((((..((((........))).)...)))))) ( -32.20) >DroEre_CAF1 150472 117 + 1 CUACCU--UUUUUUGGGUUAAAAGUUCCGGCAGGUGGGCCGUCGCUCUGUGGACUACCUUCACUUUU-UUGGUGGCUCCUAAUUUCGGGUAGAACCCGAUCUGCGCCGGAGCAGCUGCCC ......--......((((.....((((((((.((.(((((..((....(((((.....)))))....-.))..)))))))....(((((.....))))).....))))))))....)))) ( -44.50) >DroYak_CAF1 154616 117 + 1 CUGCCU--UUUUUUGGGCUAAAAGUUCCGGCAGGUGGGCCAUCGGUCUGCUGACUACCUUCACUUUU-UCGGUGGCUCCUAAUUUUGGGUAGAACCCGAUCUGCACCGGAGCAGCUGGAC ..((((--......)))).....((((((((((((((((((((((.....(((......))).....-)))))))))).......((((.....))))))))))..))))))........ ( -43.10) >consensus CUGCCU__UUUUUUGGGUUAAAAGUUCCGGCAGGUGGGCCAUCGGUCGGUGGACUACCUUCACUUUUUUCGGUGGCUCCUAAUUUUGGGUAGAACCCGAUCUGCACCGGAGCAGCUGCCC ..((((........)))).....((((((((((((((((((((((..((((((.....))))))....)))))))))).......((((.....))))))))))..))))))........ (-31.40 = -33.56 + 2.16)

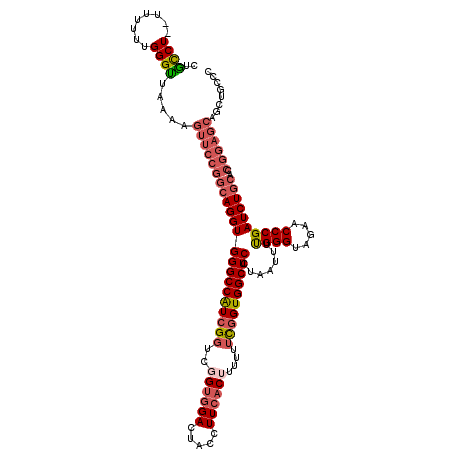
| Location | 11,415,210 – 11,415,330 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.92 |
| Mean single sequence MFE | -28.55 |
| Consensus MFE | -26.00 |
| Energy contribution | -25.72 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.23 |
| SVM RNA-class probability | 0.933805 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11415210 120 + 27905053 UGAAGAAGUAGCGCACCAGAAAACGCCUCUCAUUUUUCAACUCUGAUCAGCGAAGGUUGCCAUCUAAUUAAGUAAAUUGUCCGUUUUACAUAUGUUUCCUGGUGCCACUAAUUUUUAUGA ..(((((.(((.(((((((.(((((...................(((((((....))))..))).......(((((........)))))...))))).)))))))..))).))))).... ( -26.80) >DroSec_CAF1 162792 120 + 1 UGGAGAAGUAGCGCACCAGAAAACGCCUCUUAUUUUUCAACUCUGAUCAGCGAGGGCUGCCAUCUAAUUAAGUAAAUUGUCCGUUUUACAUAUGUUUCCUGGUGCCACUAAUUUUUAUGA ..(((((.(((.(((((((.(((((((.((..((..(((....)))..))..)))))..............(((((........)))))....)))).)))))))..))).))))).... ( -30.50) >DroSim_CAF1 155578 120 + 1 UGGAGAAGUAGCGCACCAGAAAACGCCUCUUAUUUUUCAACUCUGAUCAGCGAGGGCUACCAUCUAAUUAAGUAAAUUGUCCGUUUUACAUAUGUUUCCUGGUGCCACUAAUUUUUAUGA ..(((((.(((.(((((((.(((((((.((..((..(((....)))..))..)))))..............(((((........)))))....)))).)))))))..))).))))).... ( -30.50) >DroEre_CAF1 150589 120 + 1 CGCAGAAGUAGCGCACCAGAAAACGCCUCUUAUUUUUCAACUCUGAUCAGCGAAGGCUGCCAUCUAAUUAAGUAAAUUGUCCGUUUUACAUAUGUUUCCUGGUGCCACUAAUUUUUAUGA ((.((((((((.(((((((.(((((...................(((((((....))))..))).......(((((........)))))...))))).)))))))..)).)))))).)). ( -28.40) >DroYak_CAF1 154733 120 + 1 UGGAGAAGUAGCGCACCAGAAAACGCCUCUUAUUUUUCAACUCUGAUCAGCGAAGGCUGCCAUCUAAUUAAGUAAAUUGUGCGUUUUACAUAUGUUUCCCGGUGCCACUAAUUUUUAUGA ..(((((.(((.(((((..((((((((..((((((.........(((((((....))))..))).....))))))...).)))))))((....)).....)))))..))).))))).... ( -26.54) >consensus UGGAGAAGUAGCGCACCAGAAAACGCCUCUUAUUUUUCAACUCUGAUCAGCGAAGGCUGCCAUCUAAUUAAGUAAAUUGUCCGUUUUACAUAUGUUUCCUGGUGCCACUAAUUUUUAUGA ..(((((.(((.(((((((.((((((((.((.((..(((....)))..)).))))))..............(((((........)))))....)))).)))))))..))).))))).... (-26.00 = -25.72 + -0.28)

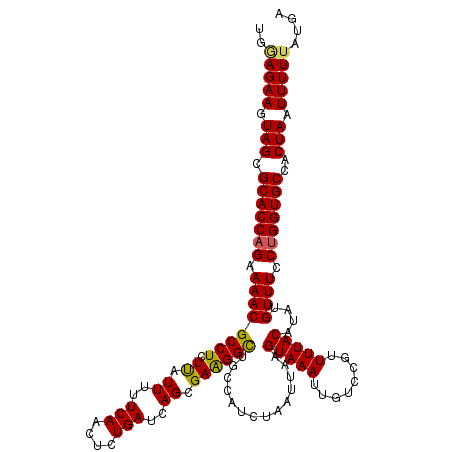
| Location | 11,415,210 – 11,415,330 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.92 |
| Mean single sequence MFE | -32.32 |
| Consensus MFE | -31.10 |
| Energy contribution | -30.98 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.96 |
| SVM decision value | 3.76 |
| SVM RNA-class probability | 0.999592 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
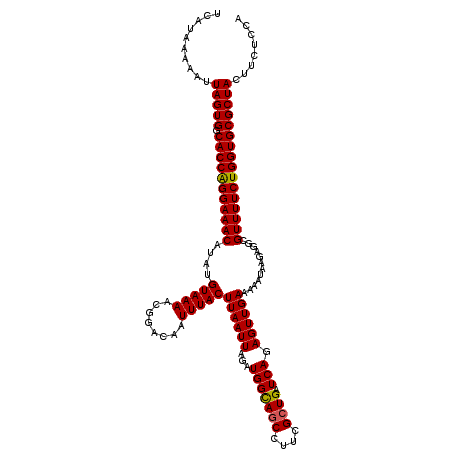
>3R_DroMel_CAF1 11415210 120 - 27905053 UCAUAAAAAUUAGUGGCACCAGGAAACAUAUGUAAAACGGACAAUUUACUUAAUUAGAUGGCAACCUUCGCUGAUCAGAGUUGAAAAAUGAGAGGCGUUUUCUGGUGCGCUACUUCUUCA ..........(((((.(((((((((((....(((((........))))).......((.(....)..))(((..(((...........)))..)))))))))))))))))))........ ( -31.40) >DroSec_CAF1 162792 120 - 1 UCAUAAAAAUUAGUGGCACCAGGAAACAUAUGUAAAACGGACAAUUUACUUAAUUAGAUGGCAGCCCUCGCUGAUCAGAGUUGAAAAAUAAGAGGCGUUUUCUGGUGCGCUACUUCUCCA ..........(((((.(((((((((((....(((((........)))))((((((...(((((((....)))).))).))))))............))))))))))))))))........ ( -33.20) >DroSim_CAF1 155578 120 - 1 UCAUAAAAAUUAGUGGCACCAGGAAACAUAUGUAAAACGGACAAUUUACUUAAUUAGAUGGUAGCCCUCGCUGAUCAGAGUUGAAAAAUAAGAGGCGUUUUCUGGUGCGCUACUUCUCCA ..........(((((.(((((((((((....(((((........)))))((((((...(((((((....))).)))).))))))............))))))))))))))))........ ( -31.30) >DroEre_CAF1 150589 120 - 1 UCAUAAAAAUUAGUGGCACCAGGAAACAUAUGUAAAACGGACAAUUUACUUAAUUAGAUGGCAGCCUUCGCUGAUCAGAGUUGAAAAAUAAGAGGCGUUUUCUGGUGCGCUACUUCUGCG ..........(((((.(((((((((((....(((((........)))))((((((...(((((((....)))).))).))))))............))))))))))))))))........ ( -33.20) >DroYak_CAF1 154733 120 - 1 UCAUAAAAAUUAGUGGCACCGGGAAACAUAUGUAAAACGCACAAUUUACUUAAUUAGAUGGCAGCCUUCGCUGAUCAGAGUUGAAAAAUAAGAGGCGUUUUCUGGUGCGCUACUUCUCCA ..........(((((.(((((((((((....(((((........)))))((((((...(((((((....)))).))).))))))............))))))))))))))))........ ( -32.50) >consensus UCAUAAAAAUUAGUGGCACCAGGAAACAUAUGUAAAACGGACAAUUUACUUAAUUAGAUGGCAGCCUUCGCUGAUCAGAGUUGAAAAAUAAGAGGCGUUUUCUGGUGCGCUACUUCUCCA ..........(((((.(((((((((((....(((((........)))))((((((...(((((((....)))).))).))))))............))))))))))))))))........ (-31.10 = -30.98 + -0.12)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:24:52 2006