| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 1,583,006 – 1,583,104 |
| Length | 98 |
| Max. P | 0.714742 |

| Location | 1,583,006 – 1,583,104 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 77.02 |
| Mean single sequence MFE | -22.85 |
| Consensus MFE | -16.46 |
| Energy contribution | -16.63 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.12 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.714742 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
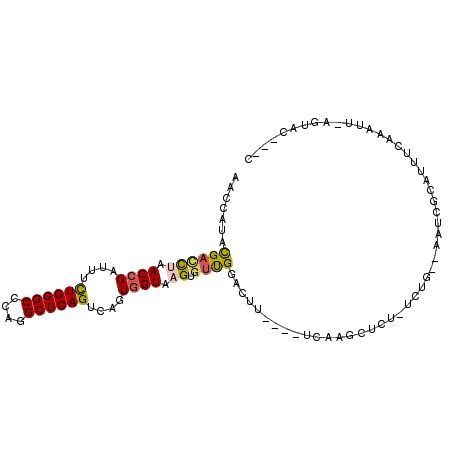
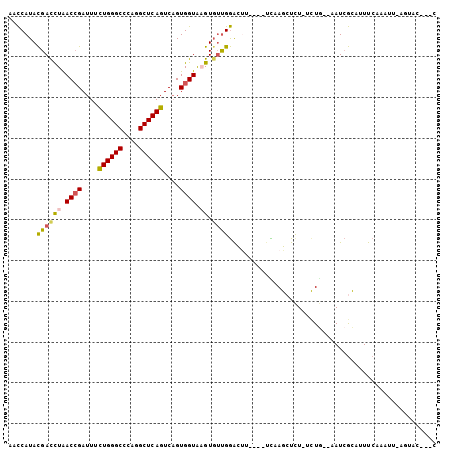
>3R_DroMel_CAF1 1583006 98 - 27905053 AACCAUACGAUCUAACCGAUUUCUGGGCCCAGGCUCAGUCAGUGGUAAGUGUUGGACUG----UCGAGCUCU-UCUG--AAUCGCAAUUCAAAUUUACUACCACC ........((((.....)))).((((((....))))))...(((((((((.(((((.((----.(((..((.-...)--).))))).)))))))))))))).... ( -26.80) >DroSec_CAF1 2138 95 - 1 AACCAUACGACCUAACCGAUUUCUGGGCCCAGGCUCAGUCAGUGGUAUGUGUUGGACUU----UCUAGAUCU-UCUG--AAUCGCAUUUCUAAUUAAGUAC---C ..(((.(((.....((((....((((((....))))))....))))...))))))((((----..((((...-....--.........))))...))))..---. ( -20.99) >DroSim_CAF1 2139 96 - 1 AACCAUACGACCUAACCGAUUUCUGGGCCCAGGCUCAGUCAGUGGUAUGUGUUGGACUU----UCAAGCUUUUUUUA--AAUCGCAUUUCUAAUUAAGUAC---C ..(((.(((.....((((....((((((....))))))....))))...))))))....----....((((..((..--((......))..))..))))..---. ( -19.50) >DroEre_CAF1 2130 95 - 1 AACCAUACGACCUAACCGAUUUCUGGGCCCAAGCUCAGUCAGUGGUAAGUGUUGGAUCU----UCAAGCUCU-UCUG--AAUCGCAUUUCCCAUUGGGUAC---C ..............(((.....((((((....)))))).((((((.((((((..(((..----(((......-..))--)))))))))).)))))))))..---. ( -23.50) >DroYak_CAF1 2130 90 - 1 AACCAUACGACCUGACCGAUUUCUGGGCCCAGGCUCAGUCAGUGGUAAGUUGUGGGUUU----UUAAGCUCU-UCUG--AAUCGCACUUCA-----GCUUU---C ((((.((((((...((((....((((((....))))))....))))..)))))))))).----.........-.(((--((......))))-----)....---. ( -27.60) >DroAna_CAF1 2121 94 - 1 AAUCAUAUGACUUGACAGAUUUUUGGGCCCAAGCUCAGUCUGUGGUGAGUGUCGAACACCUACAUAAGAUCU-UCUAUUCAUAAUAAUUCAA-------AC---C .....(((((...((.((((((((((((....)))))...(((((((.........)))).))).)))))))-))...))))).........-------..---. ( -18.70) >consensus AACCAUACGACCUAACCGAUUUCUGGGCCCAGGCUCAGUCAGUGGUAAGUGUUGGACUU____UCAAGCUCU_UCUG__AAUCGCAUUUCAAAUU_AGUAC___C .......((((((.((((....((((((....))))))....)))).)).))))................................................... (-16.46 = -16.63 + 0.17)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:42:57 2006