| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
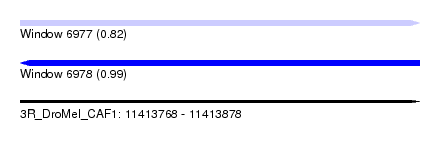
| Location | 11,413,768 – 11,413,878 |
| Length | 110 |
| Max. P | 0.988701 |

| Location | 11,413,768 – 11,413,878 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.16 |
| Mean single sequence MFE | -41.80 |
| Consensus MFE | -35.36 |
| Energy contribution | -35.52 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.823155 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11413768 110 + 27905053 AAUAAAAGGACC----------CAAGGCGAACGGAAGACAUGGCUACCUGGCCAGGACAAAGCCAGCUCCCUGUCCAUGGCGAAUCGCUGGCAGGAUGUGAGGUCCUGCCCUCGGCAGAU ......((((((----------(..(((...(....)...(((((....))))).......))).((..((((((...((((...))))))))))..))).))))))(((...))).... ( -39.30) >DroSec_CAF1 161424 110 + 1 AAUAAAAGGACC----------CAAGGCGAGCGGAUGACAUGGCUACCUGGCCAGGACAAAGCCAGCUCCCUGUCCAUGGCGAAUCGCUGACAGGAUGUGAGGUCCUGCCCUCGGCAGAU ......((((((----------((....((((((.((...(((((....)))))...))...)).))))((((((...((((...))))))))))...)).))))))(((...))).... ( -40.90) >DroSim_CAF1 152865 110 + 1 AAUAAAAGGACC----------CAAGGCGAGCGGAUGACAUGGCUACCUGGCCAGGACAAAGCCAGCUCCCUGUCCAUGGCGAAUCGCUGGCAGGAUGUGAGGUCCUGCCCUCGGCAGAU .......((((.----------..(((.((((((.((...(((((....)))))...))...)).))))))))))).((.(((......((((((((.....)))))))).))).))... ( -41.70) >DroEre_CAF1 149200 110 + 1 AAUAAAGGGACC----------CAAGGCGAGCGGAGGACAUGGCUACCUGGCCGGGACAAAGCCAGCUCCCUGUCCAUGGCGAAUCGCUGGCAAGAUGUGAGGUCCUGCCCUUGGCAGAU ......((((((----------((.(((((((.(.(((((.((((....))))(((((.......).))))))))).).))...))))).........)).))))))(((...))).... ( -38.80) >DroYak_CAF1 153250 120 + 1 AAUAAAAGGACCAAGGACCGACCAAGGCGAGCGGAAGACAGGGCUACCUGGCCAGGACAAAGCCAGCUCCCUGUCCAUGGCGAAUCGCUGGCAGGAUGUGAGGUCCUGCCCUUGGCAGAU .......((.(....).))(.(((((((((((.(..(((((((....(((((.........)))))..)))))))..).))...)))).((((((((.....)))))))))))))).... ( -48.30) >consensus AAUAAAAGGACC__________CAAGGCGAGCGGAAGACAUGGCUACCUGGCCAGGACAAAGCCAGCUCCCUGUCCAUGGCGAAUCGCUGGCAGGAUGUGAGGUCCUGCCCUCGGCAGAU ......((((((................((((((......(((((....)))))........)).))))((((((...((((...))))))))))......))))))(((...))).... (-35.36 = -35.52 + 0.16)



| Location | 11,413,768 – 11,413,878 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.16 |
| Mean single sequence MFE | -44.28 |
| Consensus MFE | -37.94 |
| Energy contribution | -37.54 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.86 |
| SVM decision value | 2.13 |
| SVM RNA-class probability | 0.988701 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11413768 110 - 27905053 AUCUGCCGAGGGCAGGACCUCACAUCCUGCCAGCGAUUCGCCAUGGACAGGGAGCUGGCUUUGUCCUGGCCAGGUAGCCAUGUCUUCCGUUCGCCUUG----------GGUCCUUUUAUU .....((((((((((((.......))))))).((((..((....(((((((.(.((((((.......)))))).)..)).)))))..)).))))))))----------)........... ( -43.40) >DroSec_CAF1 161424 110 - 1 AUCUGCCGAGGGCAGGACCUCACAUCCUGUCAGCGAUUCGCCAUGGACAGGGAGCUGGCUUUGUCCUGGCCAGGUAGCCAUGUCAUCCGCUCGCCUUG----------GGUCCUUUUAUU .....((((((((((((.......))))))).((((...((....((((((.(.((((((.......)))))).)..)).))))....))))))))))----------)........... ( -39.70) >DroSim_CAF1 152865 110 - 1 AUCUGCCGAGGGCAGGACCUCACAUCCUGCCAGCGAUUCGCCAUGGACAGGGAGCUGGCUUUGUCCUGGCCAGGUAGCCAUGUCAUCCGCUCGCCUUG----------GGUCCUUUUAUU .....((((((((((((.......))))))).((((...((....((((((.(.((((((.......)))))).)..)).))))....))))))))))----------)........... ( -42.40) >DroEre_CAF1 149200 110 - 1 AUCUGCCAAGGGCAGGACCUCACAUCUUGCCAGCGAUUCGCCAUGGACAGGGAGCUGGCUUUGUCCCGGCCAGGUAGCCAUGUCCUCCGCUCGCCUUG----------GGUCCCUUUAUU .....((((((((((((.......))))))).((((...((...(((((((.(.((((((.......)))))).)..)).)))))...))))))))))----------)........... ( -42.50) >DroYak_CAF1 153250 120 - 1 AUCUGCCAAGGGCAGGACCUCACAUCCUGCCAGCGAUUCGCCAUGGACAGGGAGCUGGCUUUGUCCUGGCCAGGUAGCCCUGUCUUCCGCUCGCCUUGGUCGGUCCUUGGUCCUUUUAUU ....(((((((((((((.......))))))).((((...((...((((((((..((((((.......))))))....))))))))...)))))))))))).((.(....).))....... ( -53.40) >consensus AUCUGCCGAGGGCAGGACCUCACAUCCUGCCAGCGAUUCGCCAUGGACAGGGAGCUGGCUUUGUCCUGGCCAGGUAGCCAUGUCAUCCGCUCGCCUUG__________GGUCCUUUUAUU ......(((((((((((.......))))))).((((...((...(((((((.(.((((((.......)))))).)..)).)))))...))))))))))...................... (-37.94 = -37.54 + -0.40)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:24:49 2006