
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 11,410,355 – 11,410,573 |
| Length | 218 |
| Max. P | 0.656818 |

| Location | 11,410,355 – 11,410,461 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.39 |
| Mean single sequence MFE | -30.72 |
| Consensus MFE | -28.14 |
| Energy contribution | -28.18 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.16 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.656818 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11410355 106 - 27905053 CGGGCAUAUCCUGUUGACAGGAGCGCAUCUGACUGCCU--------GCUGGCCUCAGGUCCUUUC-UCUUAUCUUGGCGGUUCUUUCGUCUAACAUGUGUCCUACAAAUGUUCCC----- .((((((((((((....)))))..(.((((((..(((.--------...))).)))))).)....-.........(((((.....))))).....))))))).............----- ( -30.10) >DroSec_CAF1 158003 106 - 1 CGGGCAUAUCCUGUUGACAGGAGCGCAUCUGACUGCCU--------GCUGGCCUCAGGUCCUUUC-UCUUAUCUUGGCGGUUCUUUCGUCUAACAUGUGUCCUACAAAUGUUCCC----- .((((((((((((....)))))..(.((((((..(((.--------...))).)))))).)....-.........(((((.....))))).....))))))).............----- ( -30.10) >DroSim_CAF1 149471 106 - 1 CGGGCAUAUCCUGUUGACAGGAGCGCAUCUGACUGCCU--------GCUGGCCUCAGGUCCUUUC-UCUUAUCUUGGCGGUUCUUUCGUCUAACAUGUGUCCUACAAAUGUUCCC----- .((((((((((((....)))))..(.((((((..(((.--------...))).)))))).)....-.........(((((.....))))).....))))))).............----- ( -30.10) >DroEre_CAF1 145729 107 - 1 CGGGCAUAUCCUGUUGACAGGGGCGCAUCUGACUGCCU--------GCUGGCCUCAGGUCCUUUCUUCUUAUCUUAGCGGUUCCUUCGUCUAACAUGUGUCCUACAAAUGUUGCC----- .(((((((...(((((((((((((((((((((..(((.--------...))).)))))).................)).))))))..))).))))))))))).............----- ( -29.42) >DroYak_CAF1 149604 120 - 1 CGGGCAUAUCCUGUUGACAGGAGCGCAUCUGACUGCCUGCUAUCCUGCUGGCCUCAGGUCCUUUCUUCUUAUCUUGGCGGUUCUUUCGUCUAACAUGUGUCCUACAAAUGUUCCCCCGGU ((((....(((((....)))))((((((((((..(((.((......)).))).))))..................(((((.....)))))....))))))..............)))).. ( -33.90) >consensus CGGGCAUAUCCUGUUGACAGGAGCGCAUCUGACUGCCU________GCUGGCCUCAGGUCCUUUC_UCUUAUCUUGGCGGUUCUUUCGUCUAACAUGUGUCCUACAAAUGUUCCC_____ .((((((((((((....)))))..(.((((((..(((............))).)))))).)..............(((((.....))))).....))))))).................. (-28.14 = -28.18 + 0.04)



| Location | 11,410,461 – 11,410,573 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.24 |
| Mean single sequence MFE | -34.74 |
| Consensus MFE | -25.82 |
| Energy contribution | -27.14 |
| Covariance contribution | 1.32 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.642905 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11410461 112 + 27905053 AGUGCCGAGAGCACAUUUCACACCUCACAAUGCAACAA--------UGCAACAAUGCGGCAGUGCCACAAAGGGCGGACCAGCACAGGAUAUCAGCUGCUGCCGCAGUCCCAGUUGAAAA .((((.....)))).((((((.........((((....--------))))....((((((((((((......))))...((((...........))))))))))))......).))))). ( -33.90) >DroSec_CAF1 158109 120 + 1 AGUGCCGAGAGCACAUUUCACACCUCACAAUGCAACAAUGUAAGAAUGCAACAAUGCGGCAGUGCCACAAAGGGCGGACCAGCACAGGAUAUCAGAUGCUGCCGCAGUCCCAGUUGAAAA .((((.....))))..................((((..((((....))))....((((((((((((......)))...((......)).........)))))))))......)))).... ( -32.80) >DroSim_CAF1 149577 120 + 1 AGUGCCGAGAGCACAUUUCACACCUCACAAUGCAACAAUGUGAGAAUGUAACAAUGCGGCAGUGCCACAAAGGGCGGACCAGCACAGGAUAUCAGCUGCUGCCGCAGUCCCAGUUGAAAA .((((.....)))).((((((..((((((.((...)).))))))..((..((..((((((((((((......))))...((((...........))))))))))))))..))).))))). ( -38.60) >DroEre_CAF1 145836 120 + 1 AAUGCCGAGAGCACAUUUCACACCUCACAAUGCAACAAUGCAACAAUGCACCAAUGCGGCAGUGGCACAAAGGGCGGACCUGCAGAGGAUAUCAGCUGCUGCCGCAGUCCCUGCUGAAAA ..(((.....)))..(((((..........((((....)))).....(((....((((((((..((....(((.....)))...((.....)).))..)))))))).....)))))))). ( -36.40) >DroYak_CAF1 149724 112 + 1 AGUGGCGAGAGAACAUUUCACACCUCACAAUGCAACAAUGCAACA--------AUGCGGCAGUGCCACAAAGGGCGGACCAGCACAGGAUAUCAGCUGCUGCCGCAGUCCCUGUUGAAAA .(((..((((.....)))).))).(((((.((((....))))...--------.((((((((((((......))))...((((...........)))))))))))).....)).)))... ( -32.00) >consensus AGUGCCGAGAGCACAUUUCACACCUCACAAUGCAACAAUGCAACAAUGCAACAAUGCGGCAGUGCCACAAAGGGCGGACCAGCACAGGAUAUCAGCUGCUGCCGCAGUCCCAGUUGAAAA .((((.....)))).((((((.........((((....))))............((((((((((((......)))...((......)).........)))))))))......).))))). (-25.82 = -27.14 + 1.32)

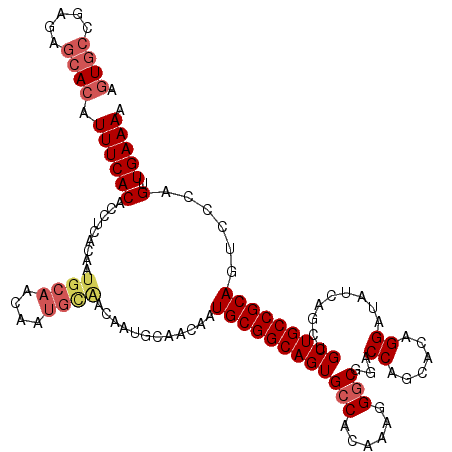

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:24:44 2006