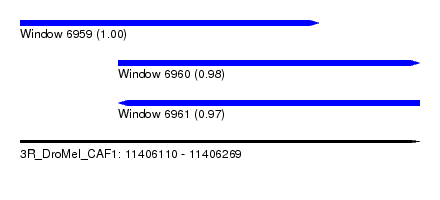
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 11,406,110 – 11,406,269 |
| Length | 159 |
| Max. P | 0.996205 |

| Location | 11,406,110 – 11,406,229 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.37 |
| Mean single sequence MFE | -31.40 |
| Consensus MFE | -28.66 |
| Energy contribution | -29.46 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.91 |
| SVM decision value | 2.67 |
| SVM RNA-class probability | 0.996205 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11406110 119 + 27905053 CUCC-GUGUUUCCCACACCAGUUGUGGAAAUCUCUGAGUUGCCUGAUUCGGAAAUUUGCGAAUUUUGAAUUUUGAUUAGAGAUUAAAUUGUCUCUGGCGAUUAGCAUCAAAUCGUUCGCU ....-.......(((((.....)))))((((.((((((((....)))))))).))))(((((((((((........(((((((......)))))))((.....)).)))))..)))))). ( -33.40) >DroSec_CAF1 153779 119 + 1 CUCC-GUGUUUCCCCCACCACUAGUGGAAAUCUCUGAGUUGCCUGAUUCGGAAAUUUGCGAAUUUUGAAUUUUGAUUAGAGAUUAAAUUGUCUCUGGCGAUUAGCAUCAAAUCGUUCGCU ....-.........((((.....))))((((.((((((((....)))))))).))))(((((((((((........(((((((......)))))))((.....)).)))))..)))))). ( -32.10) >DroSim_CAF1 145254 119 + 1 CUCC-GUGUUUCCCACACCAGUAGUGGAAAUCUCUGAGUUGCCUGAUUCGGAAAUUUGCGAAUUUUGAAUUUUGAUUAGAGAUUAAAUUGUCUCUGGCGAUUAGCAUCAAAUCGUUCACU ...(-((.....((((.......))))((((.((((((((....)))))))).))))))).....((((((((((((((((((......)))))))((.....))))))))..))))).. ( -30.10) >DroEre_CAF1 141592 112 + 1 --------CUACCCACUCCAGUAGUGGAAAUCUCCGAGUUGCCCGCUUCGGAAAUUUGCGAAUUUUGAAUUUUGAUUAGAGAUUAAAUUGUCUCUGGCGAUUAGCAUCAACUCGUUCGCU --------....(((((.....)))))((((.((((((........)))))).))))((((((.((((........(((((((......)))))))((.....)).))))...)))))). ( -31.10) >DroYak_CAF1 145247 120 + 1 CUCCCACACUUCCCACUCCAGUAGUCGAAAUCUCCGAGUUGCCUGACUCGGAAAUUUGCGAAUUUUGAAUUUUGAUUAGAGAUUAAAUUGUCUCUGGCGAUUAGCAUCCAAUCGUUCGCU ...........................((((.((((((((....)))))))).))))((((((.(((.........(((((((......)))))))((.....))...)))..)))))). ( -30.30) >consensus CUCC_GUGUUUCCCACACCAGUAGUGGAAAUCUCUGAGUUGCCUGAUUCGGAAAUUUGCGAAUUUUGAAUUUUGAUUAGAGAUUAAAUUGUCUCUGGCGAUUAGCAUCAAAUCGUUCGCU ............((((.......))))((((.((((((((....)))))))).))))(((((((((((........(((((((......)))))))((.....)).)))))..)))))). (-28.66 = -29.46 + 0.80)



| Location | 11,406,149 – 11,406,269 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.00 |
| Mean single sequence MFE | -33.66 |
| Consensus MFE | -31.26 |
| Energy contribution | -31.86 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.75 |
| SVM RNA-class probability | 0.975290 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11406149 120 + 27905053 GCCUGAUUCGGAAAUUUGCGAAUUUUGAAUUUUGAUUAGAGAUUAAAUUGUCUCUGGCGAUUAGCAUCAAAUCGUUCGCUCUGCUCGCUGAGCACUGUGCAAACAAGCGUAUGUUUGCAU ....((((((........))))))..(((((((((((((((((......)))))))((.....))))))))..))))((((........))))...(((((((((......))))))))) ( -34.00) >DroSec_CAF1 153818 120 + 1 GCCUGAUUCGGAAAUUUGCGAAUUUUGAAUUUUGAUUAGAGAUUAAAUUGUCUCUGGCGAUUAGCAUCAAAUCGUUCGCUCUGCUCGCUGAGCACUGUGCAAACAAGCGUAUGUUUGCAU ....((((((........))))))..(((((((((((((((((......)))))))((.....))))))))..))))((((........))))...(((((((((......))))))))) ( -34.00) >DroSim_CAF1 145293 120 + 1 GCCUGAUUCGGAAAUUUGCGAAUUUUGAAUUUUGAUUAGAGAUUAAAUUGUCUCUGGCGAUUAGCAUCAAAUCGUUCACUCUGCUCGCUGAGCACUGUGCAAACAAGCGUAUGUUUGCAU ....((((((........)))))).((((((((((((((((((......)))))))((.....))))))))..)))))...(((((...)))))..(((((((((......))))))))) ( -32.70) >DroEre_CAF1 141624 120 + 1 GCCCGCUUCGGAAAUUUGCGAAUUUUGAAUUUUGAUUAGAGAUUAAAUUGUCUCUGGCGAUUAGCAUCAACUCGUUCGCUCUGCUCGCUGAGCACUGUGCAAACAAGCGUAUGUUUGCAU ....(((.(((......((((((.((((........(((((((......)))))))((.....)).))))...))))))........))))))...(((((((((......))))))))) ( -34.04) >DroYak_CAF1 145287 120 + 1 GCCUGACUCGGAAAUUUGCGAAUUUUGAAUUUUGAUUAGAGAUUAAAUUGUCUCUGGCGAUUAGCAUCCAAUCGUUCGCUCUGCUCGCUGAGCACUGUGCAAACAAGCGUAUGUUUGCAU ...((.(((((......((((((.(((.........(((((((......)))))))((.....))...)))..))))))........)))))))..(((((((((......))))))))) ( -33.54) >consensus GCCUGAUUCGGAAAUUUGCGAAUUUUGAAUUUUGAUUAGAGAUUAAAUUGUCUCUGGCGAUUAGCAUCAAAUCGUUCGCUCUGCUCGCUGAGCACUGUGCAAACAAGCGUAUGUUUGCAU .((......))......(((((((((((........(((((((......)))))))((.....)).)))))..))))))..(((((...)))))..(((((((((......))))))))) (-31.26 = -31.86 + 0.60)



| Location | 11,406,149 – 11,406,269 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.00 |
| Mean single sequence MFE | -28.72 |
| Consensus MFE | -27.34 |
| Energy contribution | -27.38 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.72 |
| SVM RNA-class probability | 0.973828 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11406149 120 - 27905053 AUGCAAACAUACGCUUGUUUGCACAGUGCUCAGCGAGCAGAGCGAACGAUUUGAUGCUAAUCGCCAGAGACAAUUUAAUCUCUAAUCAAAAUUCAAAAUUCGCAAAUUUCCGAAUCAGGC .((((((((......))))))))...(((((...)))))..((((((((((.......)))))..(((((........)))))...............)))))......((......)). ( -28.40) >DroSec_CAF1 153818 120 - 1 AUGCAAACAUACGCUUGUUUGCACAGUGCUCAGCGAGCAGAGCGAACGAUUUGAUGCUAAUCGCCAGAGACAAUUUAAUCUCUAAUCAAAAUUCAAAAUUCGCAAAUUUCCGAAUCAGGC .((((((((......))))))))...(((((...)))))..((((((((((.......)))))..(((((........)))))...............)))))......((......)). ( -28.40) >DroSim_CAF1 145293 120 - 1 AUGCAAACAUACGCUUGUUUGCACAGUGCUCAGCGAGCAGAGUGAACGAUUUGAUGCUAAUCGCCAGAGACAAUUUAAUCUCUAAUCAAAAUUCAAAAUUCGCAAAUUUCCGAAUCAGGC .((((((((......))))))))...(((((...)))))..((((((((((.......)))))..(((((........)))))...............)))))......((......)). ( -26.50) >DroEre_CAF1 141624 120 - 1 AUGCAAACAUACGCUUGUUUGCACAGUGCUCAGCGAGCAGAGCGAACGAGUUGAUGCUAAUCGCCAGAGACAAUUUAAUCUCUAAUCAAAAUUCAAAAUUCGCAAAUUUCCGAAGCGGGC .((((((((......))))))))...(((((...)))))..(((((.((((((((....)))...(((((........)))))......)))))....))))).....((((...)))). ( -29.40) >DroYak_CAF1 145287 120 - 1 AUGCAAACAUACGCUUGUUUGCACAGUGCUCAGCGAGCAGAGCGAACGAUUGGAUGCUAAUCGCCAGAGACAAUUUAAUCUCUAAUCAAAAUUCAAAAUUCGCAAAUUUCCGAGUCAGGC .((((((((......))))))))...(((((...)))))..((((((((((((...)))))))..(((((........)))))...............)))))......((......)). ( -30.90) >consensus AUGCAAACAUACGCUUGUUUGCACAGUGCUCAGCGAGCAGAGCGAACGAUUUGAUGCUAAUCGCCAGAGACAAUUUAAUCUCUAAUCAAAAUUCAAAAUUCGCAAAUUUCCGAAUCAGGC .((((((((......))))))))...(((((...)))))..((((((((((.......)))))..(((((........)))))...............)))))......((......)). (-27.34 = -27.38 + 0.04)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:24:32 2006