
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 11,405,561 – 11,405,713 |
| Length | 152 |
| Max. P | 0.897322 |

| Location | 11,405,561 – 11,405,673 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.80 |
| Mean single sequence MFE | -30.19 |
| Consensus MFE | -23.69 |
| Energy contribution | -24.73 |
| Covariance contribution | 1.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.98 |
| SVM RNA-class probability | 0.893798 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
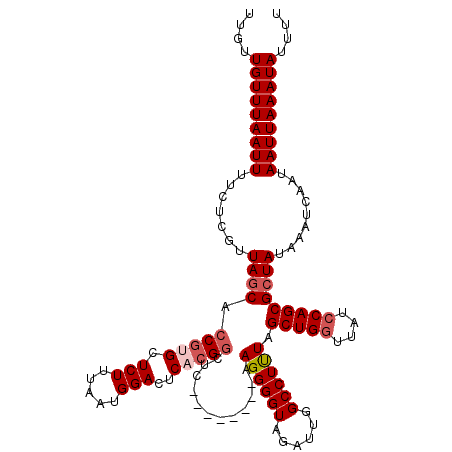
>3R_DroMel_CAF1 11405561 112 + 27905053 UUGUUGUUUAAUUUUCUCGUUAGCACCGUGCUCUUUAAUGGACUCACGGCUC--------AAGGGGUAGAUUGGCCUUUAGCUGGUUAUCCAGCGCUAUAAAUCAAUAAUUAAAUAUUUU ....(((((((((.......((((.(((((.(((.....)))..)))))...--------.((((((......)))))).(((((....))))))))).........))))))))).... ( -29.49) >DroSec_CAF1 153219 112 + 1 UUCUUGUUUAAUUUUCUCGUUAGCACCGUGCUCUUUAAUGGACUCACGGCUC--------AAGGGGUAGAUUGGCCUCUAGCUGGUUAUCCAGCGCUAUAAAUCAAUAAUUAAAUAUUUU ....(((((((((.......((((.(((((.(((.....)))..)))))...--------.((((((......)))))).(((((....))))))))).........))))))))).... ( -31.99) >DroSim_CAF1 144696 112 + 1 UUCUUGUUUAAUUUUCUCGUUAGCACCGUGCUCUUUAAUGGACUCACGGCUC--------AAGGGGUAGAUUGGCCUUUAGCUGGUUAUCCAGCGCUAUAAAUCAAUAAUUAAAUAUUUU ....(((((((((.......((((.(((((.(((.....)))..)))))...--------.((((((......)))))).(((((....))))))))).........))))))))).... ( -29.49) >DroEre_CAF1 141074 112 + 1 UUGCUGUUUAAUUUCCUCGUUAGCAGCGAGCUCUUUAAUGGACUCACGGCUG--------AAGGGGUAGGUUGGCCUUUGGCUGGUUGUGCAGCGCUAUAAAUCAAUAAUUAAAUAUUUC ..(((((.(((((.((((.(((((.(.(((.(((.....)))))).).))))--------).))))......((((...))))))))).))))).......................... ( -29.50) >DroYak_CAF1 144657 120 + 1 UGGUUGUUUAAUUUUCUCGUUAACACCGAGCUCUUUAAUGGACUCCGGGCCGAGGGACUCAAGGGGUAGAUUGGCCUUUAGCUGGUUAUCCAGCGCUAUAAAUCAAUAAUUAAAUAUUUU ....(((((((((......(((.....(((.(((.....)))))).(((((((...(((.....)))...)))))))...(((((....)))))....)))......))))))))).... ( -30.50) >consensus UUGUUGUUUAAUUUUCUCGUUAGCACCGUGCUCUUUAAUGGACUCACGGCUC________AAGGGGUAGAUUGGCCUUUAGCUGGUUAUCCAGCGCUAUAAAUCAAUAAUUAAAUAUUUU ....(((((((((.......((((.(((((.(((.....)))..)))))............((((((......)))))).(((((....))))))))).........))))))))).... (-23.69 = -24.73 + 1.04)


| Location | 11,405,601 – 11,405,713 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.58 |
| Mean single sequence MFE | -27.32 |
| Consensus MFE | -25.20 |
| Energy contribution | -25.64 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.897322 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11405601 112 - 27905053 AGGAUUUUCACACGUUCCAGUGAAUCGACCACCGUCGACCAAAAUAUUUAAUUAUUGAUUUAUAGCGCUGGAUAACCAGCUAAAGGCCAAUCUACCCCUU--------GAGCCGUGAGUC .((...(((((........)))))(((((....)))))))................(((((((...(((((....)))))....((((((........))--------).)))))))))) ( -28.70) >DroSec_CAF1 153259 112 - 1 AGGAUUUUCACACGUUCCAGUGAAUCGACCACCGUCGACCAAAAUAUUUAAUUAUUGAUUUAUAGCGCUGGAUAACCAGCUAGAGGCCAAUCUACCCCUU--------GAGCCGUGAGUC .((...(((((........)))))(((((....)))))))................(((((((...(((((....)))))....((((((........))--------).)))))))))) ( -28.70) >DroSim_CAF1 144736 112 - 1 AGGAUUUUCACACGUUCCAGUGAAUCGACCACCGUCGACCAAAAUAUUUAAUUAUUGAUUUAUAGCGCUGGAUAACCAGCUAAAGGCCAAUCUACCCCUU--------GAGCCGUGAGUC .((...(((((........)))))(((((....)))))))................(((((((...(((((....)))))....((((((........))--------).)))))))))) ( -28.70) >DroEre_CAF1 141114 112 - 1 AGGAUUUUCACACGUUCCAGUGAAUCGACCACCGUCGACCGAAAUAUUUAAUUAUUGAUUUAUAGCGCUGCACAACCAGCCAAAGGCCAACCUACCCCUU--------CAGCCGUGAGUC .((...(((((........)))))(((((....)))))))................((((((..((((((......))))...(((....))).......--------..))..)))))) ( -24.30) >DroYak_CAF1 144697 120 - 1 AGGAUUUUCACACGUUCCAGUGAAUCGACCACCGUCGACCAAAAUAUUUAAUUAUUGAUUUAUAGCGCUGGAUAACCAGCUAAAGGCCAAUCUACCCCUUGAGUCCCUCGGCCCGGAGUC .((...(((((........)))))(((((....)))))))................(((((.....(((((....)))))....((((..((........))(.....)))))..))))) ( -26.20) >consensus AGGAUUUUCACACGUUCCAGUGAAUCGACCACCGUCGACCAAAAUAUUUAAUUAUUGAUUUAUAGCGCUGGAUAACCAGCUAAAGGCCAAUCUACCCCUU________GAGCCGUGAGUC .((...(((((........)))))(((((....)))))))................(((((((.(((((((....)))))..((((..........))))..........)).))))))) (-25.20 = -25.64 + 0.44)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:24:29 2006