
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 11,405,046 – 11,405,154 |
| Length | 108 |
| Max. P | 0.898466 |

| Location | 11,405,046 – 11,405,154 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.53 |
| Mean single sequence MFE | -36.80 |
| Consensus MFE | -28.50 |
| Energy contribution | -29.70 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.77 |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.898466 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
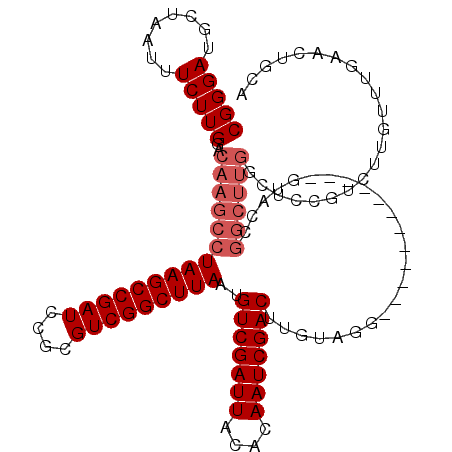
>3R_DroMel_CAF1 11405046 108 + 27905053 CGGGAUGUUAAUUUCUUGGACAAGCCUAAGCCGAUCCGCGUCGGCUUAAUGUCGAUUACACAAUCGACUUGUAGG------------GAACCGGCUUGCCUCCGUCUUGUUUGAACUGCA (((((((..........((.(((((((((((((((....)))))))))..(((((((....))))))).......------------.....))))))))..)))))))........... ( -35.80) >DroSec_CAF1 152697 108 + 1 CGGGAUGCUAAUUUCUUGGACAAGCCUAAGCCGAUCCGCGUCGGCUUAAUGUCGAUUACACAAUCGACUUGUAGG------------GUACCGGCUUGGUUCCGUCUUAUUUGAACUGCA ((((((.((((....)))).(((((((((((((((....)))))))))..(((((((....))))))).......------------.....))))))))))))................ ( -36.40) >DroSim_CAF1 144174 108 + 1 CGGGAUGCUAAUUUCUUGGACAAGCCUAAGCCGAUCCGCGUCGGCUUAAUGUCGAUUACACAAUCGACUUGUAGG------------GUACCGGCUUGGCUCCGUCUUGUUUGAACUGCA .(..((....))..)(..((((((.(..((((((.(((.((.(.((((..(((((((....)))))))...))))------------.)))))).))))))..).))))))..)...... ( -36.70) >DroEre_CAF1 140655 89 + 1 CGGGAUGCUAAUUUCUUGGACAAGCCUAAGCCGAUCCACGUCGGCUUAAUGUCGAUUACACAAUCGACUGGCUGGC-------------------------------UGUUUGAACUGCA .(..((....))..)(..((((.((((((((((((....)))))))))..(((((((....))))))).....)))-------------------------------))))..)...... ( -31.80) >DroYak_CAF1 144079 120 + 1 CGGGAUGCUAAUUUCUUGGACAAGCCUAAGCCGAUCCACGUCGGCUUAAUGUCGAUUACACAAUCGACUGGCAGGCUACGAGCUUGGGUGCCGGCUUGGCUCCGACUUGUUUGAACUGCA .((..((((((....)))).))..))(((((((((....)))))))))..(((((((....)))))))..((((...(((((.(((((.(((.....))))))))))))).....)))). ( -43.30) >consensus CGGGAUGCUAAUUUCUUGGACAAGCCUAAGCCGAUCCGCGUCGGCUUAAUGUCGAUUACACAAUCGACUUGUAGG____________GUACCGGCUUGGCUCCGUCUUGUUUGAACUGCA (((((........)))))..(((((((((((((((....)))))))))..(((((((....)))))))........................))))))...................... (-28.50 = -29.70 + 1.20)


| Location | 11,405,046 – 11,405,154 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.53 |
| Mean single sequence MFE | -34.74 |
| Consensus MFE | -26.46 |
| Energy contribution | -28.46 |
| Covariance contribution | 2.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.81 |
| SVM RNA-class probability | 0.856735 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11405046 108 - 27905053 UGCAGUUCAAACAAGACGGAGGCAAGCCGGUUC------------CCUACAAGUCGAUUGUGUAAUCGACAUUAAGCCGACGCGGAUCGGCUUAGGCUUGUCCAAGAAAUUAACAUCCCG ................(((.((((((((.....------------.......(((((((....)))))))..((((((((......)))))))))))))))).((....))......))) ( -34.90) >DroSec_CAF1 152697 108 - 1 UGCAGUUCAAAUAAGACGGAACCAAGCCGGUAC------------CCUACAAGUCGAUUGUGUAAUCGACAUUAAGCCGACGCGGAUCGGCUUAGGCUUGUCCAAGAAAUUAGCAUCCCG (((.(((.......)))(((..((((((.....------------.......(((((((....)))))))..((((((((......))))))))))))))))).........)))..... ( -35.00) >DroSim_CAF1 144174 108 - 1 UGCAGUUCAAACAAGACGGAGCCAAGCCGGUAC------------CCUACAAGUCGAUUGUGUAAUCGACAUUAAGCCGACGCGGAUCGGCUUAGGCUUGUCCAAGAAAUUAGCAUCCCG (((...........((((.((((((((((((.(------------(......(((((((....))))))).....((....)))))))))))).)))))))).((....)).)))..... ( -35.30) >DroEre_CAF1 140655 89 - 1 UGCAGUUCAAACA-------------------------------GCCAGCCAGUCGAUUGUGUAAUCGACAUUAAGCCGACGUGGAUCGGCUUAGGCUUGUCCAAGAAAUUAGCAUCCCG (((((((......-------------------------------((.((((.(((((((....)))))))..((((((((......)))))))))))).))......)))).)))..... ( -29.10) >DroYak_CAF1 144079 120 - 1 UGCAGUUCAAACAAGUCGGAGCCAAGCCGGCACCCAAGCUCGUAGCCUGCCAGUCGAUUGUGUAAUCGACAUUAAGCCGACGUGGAUCGGCUUAGGCUUGUCCAAGAAAUUAGCAUCCCG (((..............(((..((((((((((.....((.....)).)))).(((((((....)))))))..((((((((......)))))))))))))))))((....)).)))..... ( -39.40) >consensus UGCAGUUCAAACAAGACGGAGCCAAGCCGGUAC____________CCUACAAGUCGAUUGUGUAAUCGACAUUAAGCCGACGCGGAUCGGCUUAGGCUUGUCCAAGAAAUUAGCAUCCCG (((..............(((..((((((........................(((((((....)))))))..((((((((......)))))))))))))))))((....)).)))..... (-26.46 = -28.46 + 2.00)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:24:27 2006