
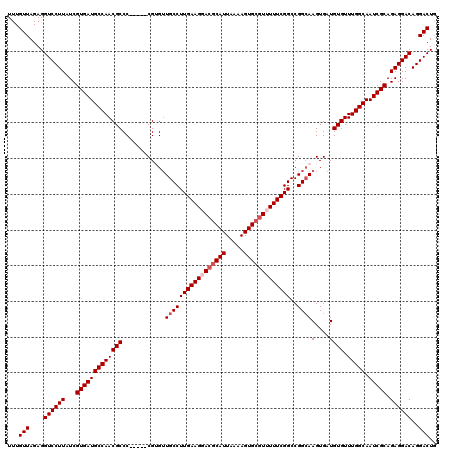
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 11,403,796 – 11,404,010 |
| Length | 214 |
| Max. P | 0.999671 |

| Location | 11,403,796 – 11,403,911 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.47 |
| Mean single sequence MFE | -43.28 |
| Consensus MFE | -37.27 |
| Energy contribution | -38.47 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.86 |
| SVM decision value | 2.55 |
| SVM RNA-class probability | 0.995173 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11403796 115 + 27905053 UUUGUUAGAGGUCCUUAUCGUGAUGCCAACGCCC-----CGUGUUGCCUUGAAGGACGCAUUAAAAGUGCGUUUUUCGGCCGGCAAGUGAUGUGUUUGGCAAUCGCAGAGGACAGGACUG ...(((....((((((...(((((((((((((..-----....((((((((((((((((((.....)))))))))))))..))))).....))).))))).))))).))))))..))).. ( -45.50) >DroSec_CAF1 151437 115 + 1 UUUGUUAGAGGUCCUUAUCGUGAUGCCAACGCCC-----CGUGUUGCCUUGAAGGACGCAUUAAAAGUGCGUUUUUCGGCCGGCAAGUGAUGUGUUUGGCAAUCGCAGAGGACAGGACUG ...(((....((((((...(((((((((((((..-----....((((((((((((((((((.....)))))))))))))..))))).....))).))))).))))).))))))..))).. ( -45.50) >DroSim_CAF1 142912 115 + 1 UUUGUUAGAGGUCCUUAUCGUGAUGCCAACGCCC-----CGUGUUGCCUUGAAGGACACAUUAAAAGUGCGUUUUUCGGCCGGCAAGUGAUGUGUUUGGCAAUCGCAGAGGACAGGACUG ...(((....((((((...(((((((((((((..-----....((((((((((((((.(((.....))).)))))))))..))))).....))).))))).))))).))))))..))).. ( -40.10) >DroEre_CAF1 139503 115 + 1 UUUGUUAGAGGUCCUUAUCGUGAUGCCAACGCCC-----CGAGUUGCCUUGAAGUAGGCAUUAAAAGUGCGUUUUUCGGCCGGCAAGUGAUGUGUUUGGCAAUCGCAGAGGACAGGACUG ...(((....((((((...(((((((((((((..-----((..(((((((((((.(.((((.....)))).).))))))..))))).))..))).))))).))))).))))))..))).. ( -40.20) >DroYak_CAF1 142631 120 + 1 UUUGUUAGAGGUCCUUAUCGUGAUGCCAACGCCCCGCUCCGAGAUGCCUUGAAGCACGCAUUAAAAGUGCGUUUUUCGGCCGGGAAGUGAUGUGUUUGGCAAUCGCAGAGGACAGGACUC ...(((....((((((...(((((((((((((..(((((((....(((..((((.((((((.....)))))).))))))))))..))))..))).))))).))))).))))))..))).. ( -45.10) >consensus UUUGUUAGAGGUCCUUAUCGUGAUGCCAACGCCC_____CGUGUUGCCUUGAAGGACGCAUUAAAAGUGCGUUUUUCGGCCGGCAAGUGAUGUGUUUGGCAAUCGCAGAGGACAGGACUG ...(((....((((((...(((((((((((((...........((((((((((((((((((.....)))))))))))))..))))).....))).))))).))))).))))))..))).. (-37.27 = -38.47 + 1.20)


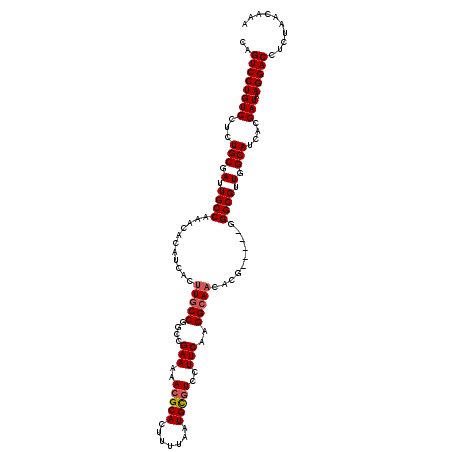
| Location | 11,403,796 – 11,403,911 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.47 |
| Mean single sequence MFE | -31.90 |
| Consensus MFE | -28.28 |
| Energy contribution | -28.72 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.795146 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11403796 115 - 27905053 CAGUCCUGUCCUCUGCGAUUGCCAAACACAUCACUUGCCGGCCGAAAAACGCACUUUUAAUGCGUCCUUCAAGGCAACACG-----GGGCGUUGGCAUCACGAUAAGGACCUCUAACAAA .......(((((...((..((((((...(.....(((((....(((..(((((.......)))))..)))..)))))....-----.)...))))))...))...))))).......... ( -31.70) >DroSec_CAF1 151437 115 - 1 CAGUCCUGUCCUCUGCGAUUGCCAAACACAUCACUUGCCGGCCGAAAAACGCACUUUUAAUGCGUCCUUCAAGGCAACACG-----GGGCGUUGGCAUCACGAUAAGGACCUCUAACAAA .......(((((...((..((((((...(.....(((((....(((..(((((.......)))))..)))..)))))....-----.)...))))))...))...))))).......... ( -31.70) >DroSim_CAF1 142912 115 - 1 CAGUCCUGUCCUCUGCGAUUGCCAAACACAUCACUUGCCGGCCGAAAAACGCACUUUUAAUGUGUCCUUCAAGGCAACACG-----GGGCGUUGGCAUCACGAUAAGGACCUCUAACAAA .......(((((...((..((((((...(.....(((((....(((..(((((.......)))))..)))..)))))....-----.)...))))))...))...))))).......... ( -29.80) >DroEre_CAF1 139503 115 - 1 CAGUCCUGUCCUCUGCGAUUGCCAAACACAUCACUUGCCGGCCGAAAAACGCACUUUUAAUGCCUACUUCAAGGCAACUCG-----GGGCGUUGGCAUCACGAUAAGGACCUCUAACAAA .......(((((...((..((((((..........((((..((((((((.....))))..(((((......)))))..)))-----)))))))))))...))...))))).......... ( -28.80) >DroYak_CAF1 142631 120 - 1 GAGUCCUGUCCUCUGCGAUUGCCAAACACAUCACUUCCCGGCCGAAAAACGCACUUUUAAUGCGUGCUUCAAGGCAUCUCGGAGCGGGGCGUUGGCAUCACGAUAAGGACCUCUAACAAA (((....(((((...((..(((((........((.(((((.((((...(((((.......)))))(((....)))...))))..))))).)))))))...))...))))))))....... ( -37.50) >consensus CAGUCCUGUCCUCUGCGAUUGCCAAACACAUCACUUGCCGGCCGAAAAACGCACUUUUAAUGCGUCCUUCAAGGCAACACG_____GGGCGUUGGCAUCACGAUAAGGACCUCUAACAAA ..((((((((...(((.(.((((...........(((((....(((..(((((.......)))))..)))..)))))..........)))).).)))....))).))))).......... (-28.28 = -28.72 + 0.44)

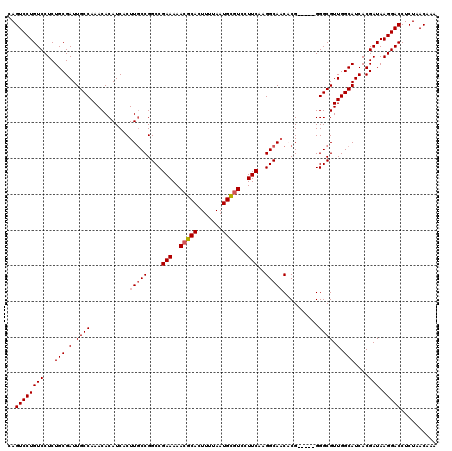
| Location | 11,403,831 – 11,403,951 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.25 |
| Mean single sequence MFE | -42.70 |
| Consensus MFE | -38.34 |
| Energy contribution | -39.42 |
| Covariance contribution | 1.08 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.66 |
| SVM RNA-class probability | 0.970433 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11403831 120 + 27905053 GUGUUGCCUUGAAGGACGCAUUAAAAGUGCGUUUUUCGGCCGGCAAGUGAUGUGUUUGGCAAUCGCAGAGGACAGGACUGGCUAAACACACAAAUAUCCUGCCGCUGCUCGGAUGCACGC ((((..((..(((((((((((.....)))))))))))(((.((....((.((((((((((..((.(........)))...)))))))))))).....)).))).......))..)))).. ( -43.30) >DroSec_CAF1 151472 120 + 1 GUGUUGCCUUGAAGGACGCAUUAAAAGUGCGUUUUUCGGCCGGCAAGUGAUGUGUUUGGCAAUCGCAGAGGACAGGACUGGCCAAACACACAAAUAUCCUGCCGCUGCUCGGAUGCACGC ((((..((..(((((((((((.....)))))))))))(((.((....((.((((((((((..((.(........)))...)))))))))))).....)).))).......))..)))).. ( -45.60) >DroSim_CAF1 142947 120 + 1 GUGUUGCCUUGAAGGACACAUUAAAAGUGCGUUUUUCGGCCGGCAAGUGAUGUGUUUGGCAAUCGCAGAGGACAGGACUGGCCAAACACACAAAUAUCCUGCCGCUGCUCGGAUGCACGC ((((..((.....)))))).......((((((((..((((.((((..((.((((((((((..((.(........)))...)))))))))))).......))))))))...)))))))).. ( -40.20) >DroEre_CAF1 139538 120 + 1 GAGUUGCCUUGAAGUAGGCAUUAAAAGUGCGUUUUUCGGCCGGCAAGUGAUGUGUUUGGCAAUCGCAGAGGACAGGACUGGCCAAACACACAAAUAUCCUGCCGCUGCUCGGAUGCACAC ....(((((......)))))......((((((((..((((.((((..((.((((((((((..((.(........)))...)))))))))))).......))))))))...)))))))).. ( -42.50) >DroYak_CAF1 142671 120 + 1 GAGAUGCCUUGAAGCACGCAUUAAAAGUGCGUUUUUCGGCCGGGAAGUGAUGUGUUUGGCAAUCGCAGAGGACAGGACUCGCCAAACACACGAAUAUCCUGCCGCUGCUCGGAUGCACGC (((..((...((((.((((((.....)))))).))))(((.((((.((..((((((((((.......(((.......)))))))))))))...)).)))))))))..))).......... ( -41.91) >consensus GUGUUGCCUUGAAGGACGCAUUAAAAGUGCGUUUUUCGGCCGGCAAGUGAUGUGUUUGGCAAUCGCAGAGGACAGGACUGGCCAAACACACAAAUAUCCUGCCGCUGCUCGGAUGCACGC ((((..((..(((((((((((.....)))))))))))(((.((....((.((((((((((..((.(........)))...)))))))))))).....)).))).......))..)))).. (-38.34 = -39.42 + 1.08)

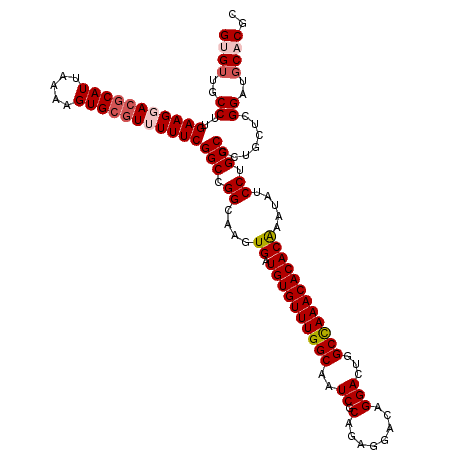
| Location | 11,403,831 – 11,403,951 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.25 |
| Mean single sequence MFE | -42.02 |
| Consensus MFE | -39.50 |
| Energy contribution | -39.78 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.94 |
| SVM decision value | 3.87 |
| SVM RNA-class probability | 0.999671 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11403831 120 - 27905053 GCGUGCAUCCGAGCAGCGGCAGGAUAUUUGUGUGUUUAGCCAGUCCUGUCCUCUGCGAUUGCCAAACACAUCACUUGCCGGCCGAAAAACGCACUUUUAAUGCGUCCUUCAAGGCAACAC ((.(((......)))))((((((......((((((((.((.(((((........).)))))).))))))))..)))))).((((((..(((((.......)))))..)))..)))..... ( -37.20) >DroSec_CAF1 151472 120 - 1 GCGUGCAUCCGAGCAGCGGCAGGAUAUUUGUGUGUUUGGCCAGUCCUGUCCUCUGCGAUUGCCAAACACAUCACUUGCCGGCCGAAAAACGCACUUUUAAUGCGUCCUUCAAGGCAACAC ((.(((......)))))((((((......(((((((((((.(((((........).)))))))))))))))..)))))).((((((..(((((.......)))))..)))..)))..... ( -43.70) >DroSim_CAF1 142947 120 - 1 GCGUGCAUCCGAGCAGCGGCAGGAUAUUUGUGUGUUUGGCCAGUCCUGUCCUCUGCGAUUGCCAAACACAUCACUUGCCGGCCGAAAAACGCACUUUUAAUGUGUCCUUCAAGGCAACAC ((.(((......)))))((((((......(((((((((((.(((((........).)))))))))))))))..)))))).((((((..(((((.......)))))..)))..)))..... ( -41.80) >DroEre_CAF1 139538 120 - 1 GUGUGCAUCCGAGCAGCGGCAGGAUAUUUGUGUGUUUGGCCAGUCCUGUCCUCUGCGAUUGCCAAACACAUCACUUGCCGGCCGAAAAACGCACUUUUAAUGCCUACUUCAAGGCAACUC (((((....((.((..(((((((......(((((((((((.(((((........).)))))))))))))))..))))))))))).....)))))......(((((......))))).... ( -43.70) >DroYak_CAF1 142671 120 - 1 GCGUGCAUCCGAGCAGCGGCAGGAUAUUCGUGUGUUUGGCGAGUCCUGUCCUCUGCGAUUGCCAAACACAUCACUUCCCGGCCGAAAAACGCACUUUUAAUGCGUGCUUCAAGGCAUCUC ..((((....((((..((((.(((.....(((((((((((.(((((........).)))))))))))))))....)))..))))....(((((.......)))))))))....))))... ( -43.70) >consensus GCGUGCAUCCGAGCAGCGGCAGGAUAUUUGUGUGUUUGGCCAGUCCUGUCCUCUGCGAUUGCCAAACACAUCACUUGCCGGCCGAAAAACGCACUUUUAAUGCGUCCUUCAAGGCAACAC ((.(((......)))))((((((......(((((((((((.(((((........).)))))))))))))))..)))))).((((((..(((((.......)))))..)))..)))..... (-39.50 = -39.78 + 0.28)

| Location | 11,403,871 – 11,403,970 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.64 |
| Mean single sequence MFE | -37.72 |
| Consensus MFE | -34.75 |
| Energy contribution | -35.43 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.711453 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11403871 99 - 27905053 GUGCAGCAUACA---------------------GCAUGCUGCGUGCAUCCGAGCAGCGGCAGGAUAUUUGUGUGUUUAGCCAGUCCUGUCCUCUGCGAUUGCCAAACACAUCACUUGCCG ..(((((((...---------------------..))))))).(((......))).(((((((......((((((((.((.(((((........).)))))).))))))))..))))))) ( -34.50) >DroSec_CAF1 151512 99 - 1 GUGCAGCGUGCA---------------------GCAUGCGGCGUGCAUCCGAGCAGCGGCAGGAUAUUUGUGUGUUUGGCCAGUCCUGUCCUCUGCGAUUGCCAAACACAUCACUUGCCG .(((.(.(((((---------------------((.....)).))))).)..))).(((((((......(((((((((((.(((((........).)))))))))))))))..))))))) ( -38.70) >DroSim_CAF1 142987 99 - 1 GUGCAGCAUGCA---------------------GCAUGCGGCGUGCAUCCGAGCAGCGGCAGGAUAUUUGUGUGUUUGGCCAGUCCUGUCCUCUGCGAUUGCCAAACACAUCACUUGCCG .(((.(.(((((---------------------((.....)).))))).)..))).(((((((......(((((((((((.(((((........).)))))))))))))))..))))))) ( -38.40) >DroEre_CAF1 139578 99 - 1 GUGCAUCAUGCA---------------------GCAUGCGGUGUGCAUCCGAGCAGCGGCAGGAUAUUUGUGUGUUUGGCCAGUCCUGUCCUCUGCGAUUGCCAAACACAUCACUUGCCG ((((((..(((.---------------------....)))..))))))........(((((((......(((((((((((.(((((........).)))))))))))))))..))))))) ( -37.10) >DroYak_CAF1 142711 120 - 1 GUGCAUCAUGCAGCAAGCAGCAAGCAGCAAGCAGCAUGCGGCGUGCAUCCGAGCAGCGGCAGGAUAUUCGUGUGUUUGGCGAGUCCUGUCCUCUGCGAUUGCCAAACACAUCACUUCCCG (((.(((.(((.((..((.(((.((........)).))).)).(((......))))).))).)))....(((((((((((.(((((........).))))))))))))))))))...... ( -39.90) >consensus GUGCAGCAUGCA_____________________GCAUGCGGCGUGCAUCCGAGCAGCGGCAGGAUAUUUGUGUGUUUGGCCAGUCCUGUCCUCUGCGAUUGCCAAACACAUCACUUGCCG ..((.((((((......................)))))).)).(((......))).(((((((......(((((((((((.(((((........).)))))))))))))))..))))))) (-34.75 = -35.43 + 0.68)

| Location | 11,403,911 – 11,404,010 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.39 |
| Mean single sequence MFE | -34.36 |
| Consensus MFE | -31.85 |
| Energy contribution | -32.33 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.29 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.687471 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11403911 99 - 27905053 UCUCCACUCAAGUGUUAAGUGUCAGAUGCAGGAUAAAAGCGUGCAGCAUACA---------------------GCAUGCUGCGUGCAUCCGAGCAGCGGCAGGAUAUUUGUGUGUUUAGC ....(((.((((((((...((((...(((.((((....(((((((((((...---------------------..)))))))))))))))..)))..)))).)))))))).)))...... ( -36.40) >DroSec_CAF1 151552 99 - 1 UCUCCACUCAAGUGUUAAGUGUCAGAUGCAGGAUAAAAGCGUGCAGCGUGCA---------------------GCAUGCGGCGUGCAUCCGAGCAGCGGCAGGAUAUUUGUGUGUUUGGC ....(((.((((((((...((((...(((.((((....((((((.(((((..---------------------.))))).))))))))))..)))..)))).)))))))).)))...... ( -33.10) >DroSim_CAF1 143027 99 - 1 UCUCCACUCAAGUGUUAAGUGUCAGAUGCAGGAUAAAAGCGUGCAGCAUGCA---------------------GCAUGCGGCGUGCAUCCGAGCAGCGGCAGGAUAUUUGUGUGUUUGGC ....(((.((((((((...((((...(((.((((....((((((.(((((..---------------------.))))).))))))))))..)))..)))).)))))))).)))...... ( -33.50) >DroEre_CAF1 139618 99 - 1 UCUCCACUCAAGUGUUAAGUGUCAGAUGCAGGAUAAAAGCGUGCAUCAUGCA---------------------GCAUGCGGUGUGCAUCCGAGCAGCGGCAGGAUAUUUGUGUGUUUGGC ....(((.((((((((...((((.((((((.(.......).))))))..((.---------------------((...(((.......))).)).)))))).)))))))).)))...... ( -31.20) >DroYak_CAF1 142751 120 - 1 UCUCCACUCAAGUGUUAAGUGUCAGAUGCAGGAUAAAAGCGUGCAUCAUGCAGCAAGCAGCAAGCAGCAAGCAGCAUGCGGCGUGCAUCCGAGCAGCGGCAGGAUAUUCGUGUGUUUGGC ....(((....)))......((((((((((.((.....((((((..(((((.((..((........))..)).)))))..))))))((((..((....)).))))..)).)))))))))) ( -37.60) >consensus UCUCCACUCAAGUGUUAAGUGUCAGAUGCAGGAUAAAAGCGUGCAGCAUGCA_____________________GCAUGCGGCGUGCAUCCGAGCAGCGGCAGGAUAUUUGUGUGUUUGGC ....(((.((((((((...((((...(((.((((....((((((.((((((......................)))))).))))))))))..)))..)))).)))))))).)))...... (-31.85 = -32.33 + 0.48)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:24:23 2006