
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 11,401,640 – 11,401,836 |
| Length | 196 |
| Max. P | 0.999941 |

| Location | 11,401,640 – 11,401,756 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.77 |
| Mean single sequence MFE | -23.52 |
| Consensus MFE | -23.68 |
| Energy contribution | -23.52 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.50 |
| Structure conservation index | 1.01 |
| SVM decision value | 4.71 |
| SVM RNA-class probability | 0.999941 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11401640 116 + 27905053 C----AUAUAUCUGCCCACUACCAAUUUUUAGUUUACGACUCUUCACAUUUUAAUGGCCAUCGCCACAAUCCGUUAAGAGUUGGAAUUUUAUUUCUCGUAAAAGAAAUACAAUUUGAGCA .----..........................((((.((((((((.((.......((((....))))......)).))))))))(((((.(((((((......))))))).))))))))). ( -23.52) >DroSec_CAF1 149303 119 + 1 CAUACAUAUAUCUGCCCACUACCAAUU-UUAGUUUACGACUCUUCACAUUUUAAUGGCCAUCGCCACAAUCCGUUAAGAGUUGGAAUUUUAUUUCUCGUAAAAGAAAUACAAUUUGAGCA ...........................-...((((.((((((((.((.......((((....))))......)).))))))))(((((.(((((((......))))))).))))))))). ( -23.52) >DroSim_CAF1 140372 116 + 1 C----AUAUAUCUGCCCACUACCAAUUUUUAGUUUACGACUCUUCACAUUUUAAUGGCCAUCGCCACAAUCCGUUAAGAGUUGGAAUUUUAUUUCUCGUAAAAGAAAUACAAUUUGAGCA .----..........................((((.((((((((.((.......((((....))))......)).))))))))(((((.(((((((......))))))).))))))))). ( -23.52) >DroEre_CAF1 137388 115 + 1 -----AUAUGUCUGCCCACUACCAAUUUUUAGUUUACGACUCUUCACAUUUUAAUGGCCAUCGCCACAAUCCGUUAAGAGUUGGAGUUUUAUUUCUCGUAAAAGAAAUACAAUUUGAGCA -----..........................((((.((((((((.((.......((((....))))......)).))))))))(((((.(((((((......))))))).))))))))). ( -23.52) >DroYak_CAF1 140377 114 + 1 ------UAUAUCUGCCCACUACCAAUUUUUAGUUUACGACUCUUCACAUUUUAAUGGCCAUCGCCACAAUCCGUUAAGAGUUGGAAUUUUAUUUCUCGUAAAAGAAAUACAAUUUGAGCA ------.........................((((.((((((((.((.......((((....))))......)).))))))))(((((.(((((((......))))))).))))))))). ( -23.52) >consensus C____AUAUAUCUGCCCACUACCAAUUUUUAGUUUACGACUCUUCACAUUUUAAUGGCCAUCGCCACAAUCCGUUAAGAGUUGGAAUUUUAUUUCUCGUAAAAGAAAUACAAUUUGAGCA ...............................((((.((((((((.((.......((((....))))......)).))))))))(((((.(((((((......))))))).))))))))). (-23.68 = -23.52 + -0.16)


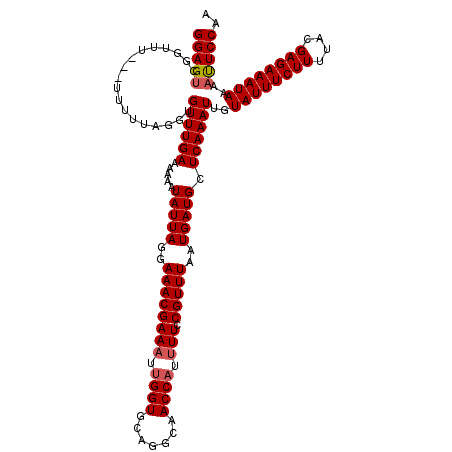
| Location | 11,401,640 – 11,401,756 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.77 |
| Mean single sequence MFE | -28.51 |
| Consensus MFE | -27.70 |
| Energy contribution | -27.70 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.97 |
| SVM decision value | 3.58 |
| SVM RNA-class probability | 0.999412 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11401640 116 - 27905053 UGCUCAAAUUGUAUUUCUUUUACGAGAAAUAAAAUUCCAACUCUUAACGGAUUGUGGCGAUGGCCAUUAAAAUGUGAAGAGUCGUAAACUAAAAAUUGGUAGUGGGCAGAUAUAU----G ((((((((((.((((((((....)))))))).))))...((((((.(((....(((((....))))).....))).)))))).....((((.....))))..)))))).......----. ( -28.30) >DroSec_CAF1 149303 119 - 1 UGCUCAAAUUGUAUUUCUUUUACGAGAAAUAAAAUUCCAACUCUUAACGGAUUGUGGCGAUGGCCAUUAAAAUGUGAAGAGUCGUAAACUAA-AAUUGGUAGUGGGCAGAUAUAUGUAUG ((((((((((.((((((((....)))))))).))))...((((((.(((....(((((....))))).....))).)))))).....((((.-...))))..))))))............ ( -28.30) >DroSim_CAF1 140372 116 - 1 UGCUCAAAUUGUAUUUCUUUUACGAGAAAUAAAAUUCCAACUCUUAACGGAUUGUGGCGAUGGCCAUUAAAAUGUGAAGAGUCGUAAACUAAAAAUUGGUAGUGGGCAGAUAUAU----G ((((((((((.((((((((....)))))))).))))...((((((.(((....(((((....))))).....))).)))))).....((((.....))))..)))))).......----. ( -28.30) >DroEre_CAF1 137388 115 - 1 UGCUCAAAUUGUAUUUCUUUUACGAGAAAUAAAACUCCAACUCUUAACGGAUUGUGGCGAUGGCCAUUAAAAUGUGAAGAGUCGUAAACUAAAAAUUGGUAGUGGGCAGACAUAU----- (((((......((((((((....))))))))..((((((((((((.(((....(((((....))))).....))).))))))..............))).)))))))).......----- ( -29.33) >DroYak_CAF1 140377 114 - 1 UGCUCAAAUUGUAUUUCUUUUACGAGAAAUAAAAUUCCAACUCUUAACGGAUUGUGGCGAUGGCCAUUAAAAUGUGAAGAGUCGUAAACUAAAAAUUGGUAGUGGGCAGAUAUA------ ((((((((((.((((((((....)))))))).))))...((((((.(((....(((((....))))).....))).)))))).....((((.....))))..))))))......------ ( -28.30) >consensus UGCUCAAAUUGUAUUUCUUUUACGAGAAAUAAAAUUCCAACUCUUAACGGAUUGUGGCGAUGGCCAUUAAAAUGUGAAGAGUCGUAAACUAAAAAUUGGUAGUGGGCAGAUAUAU____G ((((((.....((((((((....))))))))........((((((.(((....(((((....))))).....))).)))))).....((((.....))))..))))))............ (-27.70 = -27.70 + -0.00)



| Location | 11,401,676 – 11,401,796 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.17 |
| Mean single sequence MFE | -26.65 |
| Consensus MFE | -25.28 |
| Energy contribution | -24.88 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.95 |
| SVM decision value | 2.87 |
| SVM RNA-class probability | 0.997519 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11401676 120 + 27905053 UCUUCACAUUUUAAUGGCCAUCGCCACAAUCCGUUAAGAGUUGGAAUUUUAUUUCUCGUAAAAGAAAUACAAUUUGAGCAUCAUUAAACGGAAAAUGGUUGCCUGCACCAAUUUCGUUUC ...............(((....((((...(((((((((((((.(((((.(((((((......))))))).))))).))).)).)).))))))...)))).)))................. ( -26.50) >DroSec_CAF1 149342 120 + 1 UCUUCACAUUUUAAUGGCCAUCGCCACAAUCCGUUAAGAGUUGGAAUUUUAUUUCUCGUAAAAGAAAUACAAUUUGAGCAUCAUUAAACGGAAAAUGGUUGCCUGCACCAAUUUCGUUUC ...............(((....((((...(((((((((((((.(((((.(((((((......))))))).))))).))).)).)).))))))...)))).)))................. ( -26.50) >DroSim_CAF1 140408 120 + 1 UCUUCACAUUUUAAUGGCCAUCGCCACAAUCCGUUAAGAGUUGGAAUUUUAUUUCUCGUAAAAGAAAUACAAUUUGAGCAUCAUUAAACGGAAAAUGGUUGCCUGCACCAAUUUCGUUUC ...............(((....((((...(((((((((((((.(((((.(((((((......))))))).))))).))).)).)).))))))...)))).)))................. ( -26.50) >DroEre_CAF1 137423 119 + 1 UCUUCACAUUUUAAUGGCCAUCGCCACAAUCCGUUAAGAGUUGGAGUUUUAUUUCUCGUAAAAGAAAUACAAUUUGAGCAUCAUUAAACGAAA-AGGGUUCCCUGCACCAAUUUCGUUUC ((((.((.......((((....))))......)).))))(((.(((((.(((((((......))))))).))))).)))......((((((((-..(((.......)))..)))))))). ( -26.82) >DroYak_CAF1 140411 120 + 1 UCUUCACAUUUUAAUGGCCAUCGCCACAAUCCGUUAAGAGUUGGAAUUUUAUUUCUCGUAAAAGAAAUACAAUUUGAGCAUCAUUAAACGAAAAAUGGUUGCCUGCACCAAUUUCGUUUC ((((.((.......((((....))))......)).))))(((.(((((.(((((((......))))))).))))).)))......((((((((..((((.(....))))).)))))))). ( -26.92) >consensus UCUUCACAUUUUAAUGGCCAUCGCCACAAUCCGUUAAGAGUUGGAAUUUUAUUUCUCGUAAAAGAAAUACAAUUUGAGCAUCAUUAAACGGAAAAUGGUUGCCUGCACCAAUUUCGUUUC ((((.((.......((((....))))......)).))))(((.(((((.(((((((......))))))).))))).)))......((((((((...(((.......)))..)))))))). (-25.28 = -24.88 + -0.40)



| Location | 11,401,716 – 11,401,836 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.39 |
| Mean single sequence MFE | -26.38 |
| Consensus MFE | -22.88 |
| Energy contribution | -23.32 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.50 |
| SVM RNA-class probability | 0.959395 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11401716 120 - 27905053 GGAGUCGUUUUUUUUUUUUAGGUGUUUGAAAAAAUAUUAGGAAACGAAAUUGGUGCAGGCAACCAUUUUCCGUUUAAUGAUGCUCAAAUUGUAUUUCUUUUACGAGAAAUAAAAUUCCAA (((((((((...........(((((((....)))))))...((((((((.((((.......)))).))).)))))))))))..........((((((((....))))))))....))).. ( -24.70) >DroSec_CAF1 149382 117 - 1 GGAGUCGGUUU---UUUUUAGGUGUUUGAAAAAAUAUUAGGAAACGAAAUUGGUGCAGGCAACCAUUUUCCGUUUAAUGAUGCUCAAAUUGUAUUUCUUUUACGAGAAAUAAAAUUCCAA (((((((((((---(((...(((((((....))))))))))))))((((.((((.......)))).)))).......))))..........((((((((....))))))))....))).. ( -24.50) >DroSim_CAF1 140448 117 - 1 GGAGACGGUUU---UUUUUAGGUGUUUGAAAAAAUAUUAGGAAACGAAAUUGGUGCAGGCAACCAUUUUCCGUUUAAUGAUGCUCAAAUUGUAUUUCUUUUACGAGAAAUAAAAUUCCAA (((((((((((---(((...(((((((....))))))))))))))((((.((((.......)))).)))))))))...........((((.((((((((....)))))))).)))))).. ( -25.80) >DroEre_CAF1 137463 114 - 1 GGAGUCAGUUG-----UUCAGGUGUUUGAAAAAAUAUUAAGAAACGAAAUUGGUGCAGGGAACCCU-UUUCGUUUAAUGAUGCUCAAAUUGUAUUUCUUUUACGAGAAAUAAAACUCCAA (((((.(((..-----.((((((((((....)))))))...((((((((..(((.......)))..-))))))))..))).))).......((((((((....))))))))..))))).. ( -29.40) >DroYak_CAF1 140451 115 - 1 GGAGUCAGUUG-----UUCAGGUGUUUGAAAAAAUAUUAAGAAACGAAAUUGGUGCAGGCAACCAUUUUUCGUUUAAUGAUGCUCAAAUUGUAUUUCUUUUACGAGAAAUAAAAUUCCAA (((((.(((..-----.((((((((((....)))))))...((((((((.((((.......))))..))))))))..))).))).......((((((((....))))))))..))))).. ( -27.50) >consensus GGAGUCGGUUU___UUUUUAGGUGUUUGAAAAAAUAUUAGGAAACGAAAUUGGUGCAGGCAACCAUUUUCCGUUUAAUGAUGCUCAAAUUGUAUUUCUUUUACGAGAAAUAAAAUUCCAA (((((..................((((((.....(((((..((((((((.((((.......)))).))).)))))..))))).))))))..((((((((....))))))))..))))).. (-22.88 = -23.32 + 0.44)


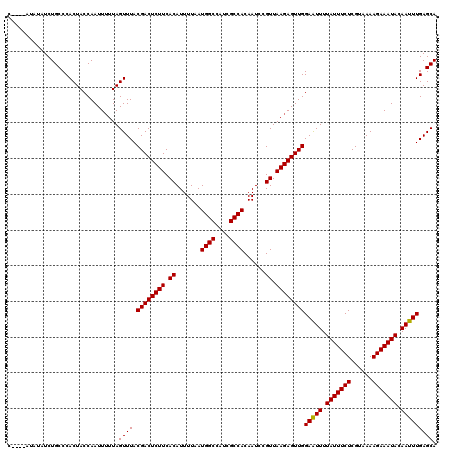
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:24:15 2006