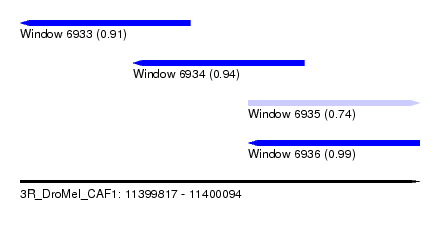
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 11,399,817 – 11,400,094 |
| Length | 277 |
| Max. P | 0.987468 |

| Location | 11,399,817 – 11,399,935 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.79 |
| Mean single sequence MFE | -40.04 |
| Consensus MFE | -25.10 |
| Energy contribution | -27.62 |
| Covariance contribution | 2.52 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.80 |
| Structure conservation index | 0.63 |
| SVM decision value | 1.05 |
| SVM RNA-class probability | 0.906938 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11399817 118 - 27905053 CAAGUGCGGCAACCUGCUGAGCUUGGCCAAUUGGGUCAGCUUAGG-GGAGUUUGGAAAAUAAAGAGGGUGUUGGAUUAGCGGCUUGGUGGUGUAAACUGCAGCUGC-AGUUGACAAGAAA (((.((((((..(((.(((((((.((((.....))))))))))))-))((((((.(..((....(((.(((((...))))).))).))..).))))))...)))))-).)))........ ( -40.70) >DroSec_CAF1 147493 119 - 1 CAAGUGCGGCAACCUGCUGAGCUUGGCCAAUUGGGUCAGCUUAGGGGGAGUUUGGAAAAUAAAGAGGGUGUUAGAUUAGGGGCUUGAUGCUGUAAGCUGCAGCUGC-AGUUGACAAUAAA (((.((((((..(((.(((((((.((((.....))))))))))).)))..............................(.((((((......)))))).).)))))-).)))........ ( -42.90) >DroSim_CAF1 138575 119 - 1 CAAGUGCGGCAACCUACUGAGCUUGGCCAAUUGGGUCAGCUUAGGGGGAGCUUGGAAAAUAAAGAGGGUGUUAGAUUAGCGGCUUGGUGGUGUAAGCUGCAGCUGC-AGUUGACAAUAAA (((.((((((..((..(((((((.((((.....)))))))))))..))((((((.(..((....(((.(((((...))))).))).))..).))))))...)))))-).)))........ ( -45.20) >DroEre_CAF1 135599 117 - 1 AAAGUGCUGCAACCUGCUGAGCUUGGCCAAUUGGGUCAGCUUGGG--GCUUUUGGUAAAUAAAGAGGGUGUUAGAUUAGCGGCUUGGUUGUGUAAGCGGCAGCUGCAAGUUGACAGCA-A ....(((((((((..((((.(((((..((((..((((.((((((.--((((((..(....)..)))))).)))....)))))))..))))..)))))..)))).....)))).)))))-. ( -42.00) >DroYak_CAF1 138545 105 - 1 CAAGUGCCGCAACCUGCUGAGCUUGGCCAAUUGGGUCAGCUUGGG-GGCUUUGGGAAAACAGGAAGGGUGUUGGAUUAGCGGCUUAGUUGUGUA-------------AGUUGACAGGG-A .....(((((..(((.(..((((.((((.....))))))))..))-))((((.(.....)..))))............)))))....((.(((.-------------.....))).))-. ( -29.40) >consensus CAAGUGCGGCAACCUGCUGAGCUUGGCCAAUUGGGUCAGCUUAGG_GGAGUUUGGAAAAUAAAGAGGGUGUUAGAUUAGCGGCUUGGUGGUGUAAGCUGCAGCUGC_AGUUGACAAGAAA (((.((((((.((((.(((((((.((((.....)))))))))))......(((........))).)))).........((((((((......)))))))).))))).).)))........ (-25.10 = -27.62 + 2.52)

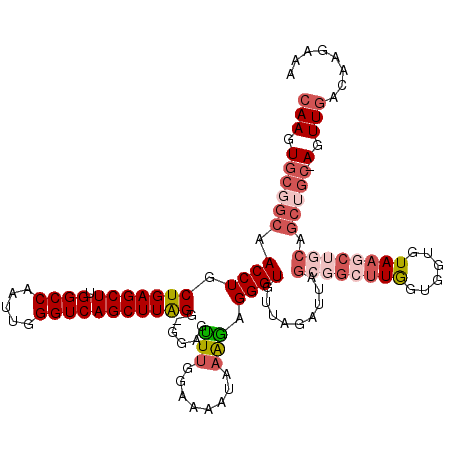
| Location | 11,399,895 – 11,400,014 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.90 |
| Mean single sequence MFE | -37.71 |
| Consensus MFE | -32.44 |
| Energy contribution | -32.36 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.26 |
| SVM RNA-class probability | 0.937850 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11399895 119 - 27905053 CCAGCAGAAGAAUCAGCCGGAAGCUGAAAGAACU-UUGCAGUGGAAAAAUAUAGUACACGAGUAUCUUGUGGAUGCGACGCAAGUGCGGCAACCUGCUGAGCUUGGCCAAUUGGGUCAGC .((((((.....((((.(....))))).......-((((.(((.............)))..(((.((((((.......))))))))).)))).)))))).(((.((((.....))))))) ( -38.32) >DroSec_CAF1 147572 120 - 1 CCAGCAGAAGAAUCAGCCGGAAGCUGAAAGAACUUUUGCAGUGGAAAAAUAUAAUACACGAGUAUCUUGUGGAUGCGACGCAAGUGCGGCAACCUGCUGAGCUUGGCCAAUUGGGUCAGC .((((((.....((((.(....)))))........((((.(((.............)))..(((.((((((.......))))))))).)))).)))))).(((.((((.....))))))) ( -38.32) >DroSim_CAF1 138654 120 - 1 CCAGCAGAAGAAUCAGCCGGAAGCUGAAAGAACUUUUGCAGUGGAAAAAUAUAGUACACGAGUAUCUUUUGGAUGCGACGCAAGUGCGGCAACCUACUGAGCUUGGCCAAUUGGGUCAGC ...(((((((..((((.(....))))).....)))))))(((((.........((((.((.(((((.....)))))..))...))))(....))))))..(((.((((.....))))))) ( -35.20) >DroEre_CAF1 135676 119 - 1 CCAGCAGAAGAAUCAGCCGGAAGCUGAAAGAAAU-GCGCAGUGGAAGAAUAUAGAACACGAGUAUCUUGUGGAUGCGGGGAAAGUGCUGCAACCUGCUGAGCUUGGCCAAUUGGGUCAGC .((((((.....((((.(....)))))......(-((((((((.............)))..(((((.....)))))........))).)))..)))))).(((.((((.....))))))) ( -36.52) >DroYak_CAF1 138610 119 - 1 CCAGCAGAAGAAUCAGCCGGCAGCUGAAAGAAAC-UCGCAGUGGAGAAAUAUAGUAUUCGAGUAUCUUGUGGAUGCGGGGCAAGUGCCGCAACCUGCUGAGCUUGGCCAAUUGGGUCAGC .((((((.....(((((.....)))))((((.((-(((.((((.(....)....))))))))).)))).....(((((.(....).)))))..)))))).(((.((((.....))))))) ( -40.20) >consensus CCAGCAGAAGAAUCAGCCGGAAGCUGAAAGAACU_UUGCAGUGGAAAAAUAUAGUACACGAGUAUCUUGUGGAUGCGACGCAAGUGCGGCAACCUGCUGAGCUUGGCCAAUUGGGUCAGC .((((((.....(((((.....)))))..........((.(((.............)))..(((((.....)))))..(((....)))))...)))))).(((.((((.....))))))) (-32.44 = -32.36 + -0.08)

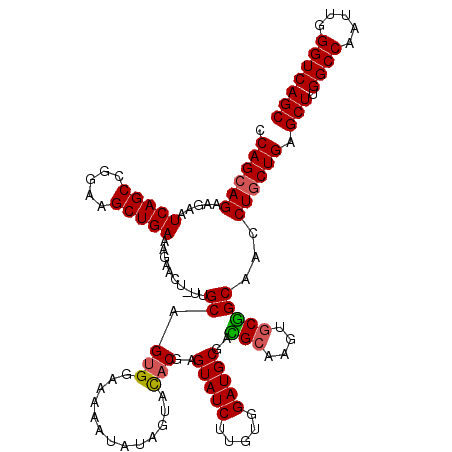

| Location | 11,399,975 – 11,400,094 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.65 |
| Mean single sequence MFE | -19.57 |
| Consensus MFE | -15.21 |
| Energy contribution | -14.97 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.737245 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11399975 119 + 27905053 UGCAA-AGUUCUUUCAGCUUCCGGCUGAUUCUUCUGCUGGCUGCUUCAAAAAUUACAUUAAACAUACUUUUAUUUAAUUCUCGCUUUCCAGCCACCCCCUUGACCAACAUCCCACUUCGG .((((-((.....(((((.....)))))..))).)))((((((.......(((((...((((......))))..))))).........)))))).......................... ( -17.49) >DroSec_CAF1 147652 120 + 1 UGCAAAAGUUCUUUCAGCUUCCGGCUGAUUCUUCUGCUGGCUGCUUCAAAAAUUACAUUAAACAUACUUUUAUUUAAUUCUCGCUUUCCAGCCACCCCCUUGACCAUCAUCCCACUUCUG .(((.(((.....(((((.....)))))..))).)))((((((.......(((((...((((......))))..))))).........)))))).......................... ( -18.69) >DroSim_CAF1 138734 120 + 1 UGCAAAAGUUCUUUCAGCUUCCGGCUGAUUCUUCUGCUGGCUGCUUCAAAAAUUACAUUAAACAUACUUUUAUUUAAUUCUCGCUUUCCAGCCACCCCCUUGACCAUCAUCCCACUUCUG .(((.(((.....(((((.....)))))..))).)))((((((.......(((((...((((......))))..))))).........)))))).......................... ( -18.69) >DroEre_CAF1 135756 119 + 1 UGCGC-AUUUCUUUCAGCUUCCGGCUGAUUCUUCUGCUGGCUGCUUCAAAAAUUACAUUAAACAUACUUUUAUUUAAUUCUCGCUCCCCAGCCACCCUCUUGACCAUCAUCCCACUUCGG ...((-(......(((((.....)))))......)))((((((.......(((((...((((......))))..))))).........)))))).......................... ( -17.29) >DroYak_CAF1 138690 116 + 1 UGCGA-GUUUCUUUCAGCUGCCGGCUGAUUCUUCUGCUGGCUGCUUCAGAAAUUACAUUAAACAUACUUUUAUUUAAUUCUCGCUUUGCAGCCACCCUCUUGACCAUCAACCCACU---G .((((-((((((...(((.((((((.((....)).)))))).)))..)))))....((((((.(......).)))))).)))))................................---. ( -25.70) >consensus UGCAA_AGUUCUUUCAGCUUCCGGCUGAUUCUUCUGCUGGCUGCUUCAAAAAUUACAUUAAACAUACUUUUAUUUAAUUCUCGCUUUCCAGCCACCCCCUUGACCAUCAUCCCACUUCGG .(((.........(((((.....)))))......)))((((((.......(((((...((((......))))..))))).........)))))).......................... (-15.21 = -14.97 + -0.24)


| Location | 11,399,975 – 11,400,094 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.65 |
| Mean single sequence MFE | -29.96 |
| Consensus MFE | -24.42 |
| Energy contribution | -24.38 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.82 |
| SVM decision value | 2.08 |
| SVM RNA-class probability | 0.987468 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11399975 119 - 27905053 CCGAAGUGGGAUGUUGGUCAAGGGGGUGGCUGGAAAGCGAGAAUUAAAUAAAAGUAUGUUUAAUGUAAUUUUUGAAGCAGCCAGCAGAAGAAUCAGCCGGAAGCUGAAAGAACU-UUGCA ((......))......(.(((((.(.((((((.....(((((((((..((((......))))...)))))))))...)))))).).......((((.(....))))).....))-)))). ( -28.90) >DroSec_CAF1 147652 120 - 1 CAGAAGUGGGAUGAUGGUCAAGGGGGUGGCUGGAAAGCGAGAAUUAAAUAAAAGUAUGUUUAAUGUAAUUUUUGAAGCAGCCAGCAGAAGAAUCAGCCGGAAGCUGAAAGAACUUUUGCA (((((((.................(.((((((.....(((((((((..((((......))))...)))))))))...)))))).).......((((.(....)))))....))))))).. ( -29.60) >DroSim_CAF1 138734 120 - 1 CAGAAGUGGGAUGAUGGUCAAGGGGGUGGCUGGAAAGCGAGAAUUAAAUAAAAGUAUGUUUAAUGUAAUUUUUGAAGCAGCCAGCAGAAGAAUCAGCCGGAAGCUGAAAGAACUUUUGCA (((((((.................(.((((((.....(((((((((..((((......))))...)))))))))...)))))).).......((((.(....)))))....))))))).. ( -29.60) >DroEre_CAF1 135756 119 - 1 CCGAAGUGGGAUGAUGGUCAAGAGGGUGGCUGGGGAGCGAGAAUUAAAUAAAAGUAUGUUUAAUGUAAUUUUUGAAGCAGCCAGCAGAAGAAUCAGCCGGAAGCUGAAAGAAAU-GCGCA ((....(((........)))...)).((((((.....(((((((((..((((......))))...)))))))))...))))))(((......((((.(....)))))......)-))... ( -27.40) >DroYak_CAF1 138690 116 - 1 C---AGUGGGUUGAUGGUCAAGAGGGUGGCUGCAAAGCGAGAAUUAAAUAAAAGUAUGUUUAAUGUAAUUUCUGAAGCAGCCAGCAGAAGAAUCAGCCGGCAGCUGAAAGAAAC-UCGCA .---.((.(((((((..((.....(.(((((((..((.((..((((((((......))))))))....)).))...))))))).).))...))))))).)).((.((.......-)))). ( -34.30) >consensus CAGAAGUGGGAUGAUGGUCAAGGGGGUGGCUGGAAAGCGAGAAUUAAAUAAAAGUAUGUUUAAUGUAAUUUUUGAAGCAGCCAGCAGAAGAAUCAGCCGGAAGCUGAAAGAACU_UUGCA .....(((................(.((((((.....(((((((((..((((......))))...)))))))))...)))))).).......(((((.....))))).........))). (-24.42 = -24.38 + -0.04)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:24:08 2006