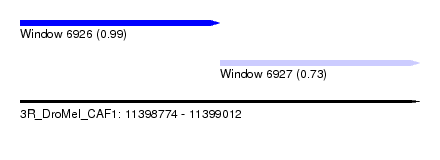
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 11,398,774 – 11,399,012 |
| Length | 238 |
| Max. P | 0.987753 |

| Location | 11,398,774 – 11,398,893 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.65 |
| Mean single sequence MFE | -32.60 |
| Consensus MFE | -20.30 |
| Energy contribution | -21.58 |
| Covariance contribution | 1.28 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.79 |
| Structure conservation index | 0.62 |
| SVM decision value | 2.09 |
| SVM RNA-class probability | 0.987753 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11398774 119 + 27905053 ACAAAACGCAUCAAAGAGGAUGAUCCCAUCCCAUCAAUAAAUCCUGAUGGCAACA-CCCCCACAUGAAAAUCUCUUCAUUGAUGGCGCGGUGUUGGGAAAUCACUUCACUGCACUAAAAU .......(((.....((((.(((((((...((((((........)))))).((((-((((((((((((......)))).)).))).).))))))))))..)))))))..)))........ ( -33.90) >DroSec_CAF1 146464 119 + 1 ACAAAACGCAUCAAAGAGGAUUAUCCCAUCCCAUCAAUAAAUCCUGAUGGCAACU-CCCCCAUAUGGAAAUCUCUUCAUUGAUGGCGCGGUGUUGGGAAAUCACUUCACUGCACUAAAAU .......(((...(((..((((.((((...((((((........)))))).(((.-((((((((((((......)))))..)))).).)).))))))))))).)))...)))........ ( -30.20) >DroSim_CAF1 137551 119 + 1 ACAAAACGCAUCAAAGAGGAUUAUCCCAUUCCAUCAAUAAAUCCUGAUGGCAACU-CCCCCACGUGGAAAUCUCUUUAUUGAUGGCGCGGUGUUGGGAAAUCACUUCACUGCACUAAAAU .......(((((((((((((((.(((....((((((........))))))..((.-.......))))))))).)))).))))).))((((((..((.......)).))))))........ ( -28.90) >DroEre_CAF1 134571 111 + 1 ---------AUCGGGGAGGAUUUUCCCCUCCCAACUAGAAAUCCUGCAGGCGGGAGGUGCCACAUUGAAAUCUCCUCAUUGAUGGCGCGCUGUUGGGAAAUCACUUCAGUGCACUAAAAU ---------...((((((....))))))(((((((.((...(((((....))))).((((((((.(((.......))).)).)))))).)))))))))...(((....)))......... ( -40.80) >DroYak_CAF1 137479 106 + 1 ACAGAACGCAUCACAGAGGAUUAUCCCAUCCCACCGGGAACUCCC--------------CCAUAUCGAAAUGUCUUCAUUGAUGGCGCGGUGUUGGGAAAUCACUUCGCUGCACUAAAAU .......(((...(..((((((.(((((...(((((((....)))--------------((((.....((((....)))).))))...)))).))))))))).))..).)))........ ( -29.20) >consensus ACAAAACGCAUCAAAGAGGAUUAUCCCAUCCCAUCAAUAAAUCCUGAUGGCAACA_CCCCCACAUGGAAAUCUCUUCAUUGAUGGCGCGGUGUUGGGAAAUCACUUCACUGCACUAAAAU ........((((((.(((((..........((((((........)))))).........((....)).....))))).))))))..((((((..((.......)).))))))........ (-20.30 = -21.58 + 1.28)


| Location | 11,398,893 – 11,399,012 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.69 |
| Mean single sequence MFE | -32.34 |
| Consensus MFE | -25.54 |
| Energy contribution | -26.06 |
| Covariance contribution | 0.52 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.727369 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11398893 119 + 27905053 CCUAUUAAAACCGUGGAUCGUCAGGGCAACAACAA-AGGCGGUGCCAAAAGCAUUAGUCAAGCAUUUUUUUUUCAGCCAUUUUUGGCUCACGGAAAAUGCAAAUUAGUCAGCCCAAACGU .((((.......))))..(((..((((........-.((((((((.....))))).)))..(((((((((....(((((....)))))...)))))))))..........))))..))). ( -32.70) >DroSec_CAF1 146583 117 + 1 CCUAUUAAAUCCGUGGAUCGGCAGGGCAACAACAAAAGGCGGUGCCAAAAGCAUUAGUCAAGCAUUUU---UUUAGCCAUUUUUGGCUCACGGAAAAUGCAAAUUAGUCAGCCCAAGCGU ..........(((.....)))..((((..........((((((((.....))))).)))..(((((((---((.(((((....)))))...)))))))))..........))))...... ( -32.90) >DroSim_CAF1 137670 118 + 1 CCUAUUAAAUCCGUGGAUCGGCAGGGCAACAACAAAAGGCGGUGCCAAAAGCAUUAGUCAAGCAUUUUU--UUUAGCCAUUUUUGGCUCACGGAAAAUGCAAAUUAGUCAGCCCAAACGU ..........(((.....)))..((((..........((((((((.....))))).)))..((((((((--(..(((((....)))))...)))))))))..........))))...... ( -33.30) >DroEre_CAF1 134682 115 + 1 CGCAUUAAGUCCCUGGAUCUGGUGGGCAGCCGCAA-AGGCGGUGCCAAAAGCAUUAGUCAAGUAUUU----UUUAGCCAUUUUCGGCUCACGGAAAAUGCAAAUUAGUCAGCCCAAACGU ..(((((..((....))..)))))((((.((((..-..))))))))....((((((((...((((((----(((((((......))))...)))))))))..))))))..))........ ( -33.00) >DroYak_CAF1 137585 115 + 1 CCUAUUAAGUCCGCGGAUCUGCAGGGCAGCAACAA-AGGCGGUGGCAAGAGCAUUAGUCAAGUAUUU----UUUAGCCAUUUUUGGCUCACGGAAAAUGCAAAUUAGUCAGCCCAAACGU ............(((....))).((((........-.(((....((....))....)))..((((((----((((((((....)))))...)))))))))..........))))...... ( -29.80) >consensus CCUAUUAAAUCCGUGGAUCGGCAGGGCAACAACAA_AGGCGGUGCCAAAAGCAUUAGUCAAGCAUUUU___UUUAGCCAUUUUUGGCUCACGGAAAAUGCAAAUUAGUCAGCCCAAACGU ........(((....))).....((((..........((((((((.....))))).)))..(((((((......(((((....))))).....)))))))..........))))...... (-25.54 = -26.06 + 0.52)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:24:00 2006