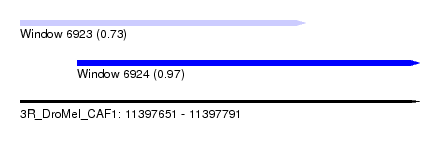
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 11,397,651 – 11,397,791 |
| Length | 140 |
| Max. P | 0.965644 |

| Location | 11,397,651 – 11,397,751 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.72 |
| Mean single sequence MFE | -31.82 |
| Consensus MFE | -27.38 |
| Energy contribution | -27.98 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.728891 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11397651 100 + 27905053 ----G----------------GAAUCUGCCGGGCGGCAGUCAUAAAGCCCAGGAAACAUGGCCAUGGUCUUCAUACCGGCUCAACACUGACAGGUGUAAAUUGCCGAGGCUUAACAAAAG ----.----------------......(((...(((((((.....((((..(....)..(((....)))........))))..(((((....)))))..))))))).))).......... ( -29.60) >DroSec_CAF1 145350 100 + 1 ----G----------------GAAUCUGCCGGUCGGCAGUCAUAAAGCCCAGGAAACAUGGCCAUGGUCUUCAUGCCGGCUCAACACUGACAGGUGUAAAUUGCCGAGGCUUAACAAAAG ----.----------------......(((..((((((((.....((((..(....)..(((.(((.....))))))))))..(((((....)))))..))))))))))).......... ( -32.20) >DroSim_CAF1 136432 100 + 1 ----G----------------GAAUCUGCCGGUCGGCAGUCAUAAAGCCCAGGAAACAUGGCCAUGGUCUUCAUGCCGGCUCAACACUGACAGGUGUAAAUUGCCGAGGCUUAACAAAAG ----.----------------......(((..((((((((.....((((..(....)..(((.(((.....))))))))))..(((((....)))))..))))))))))).......... ( -32.20) >DroEre_CAF1 133520 101 + 1 UUUCG----------------GAAUCUGCCGG---GCAGUCAUAAAGCCCAGGAAACAUGGCCAUGGUCUUCAUGCCGGCUCAACACUGACAGGUGUAAAUUGCCGAGGCUUAACAAAAG .....----------------......(((.(---(((((.....((((..(....)..(((.(((.....))))))))))..(((((....)))))..))))))..))).......... ( -29.90) >DroYak_CAF1 136308 117 + 1 UUUCGGCAUUUCAACAUUUUGGAAUCUGCCGG---GCAGUCAUAAAGCCCAGGAAACAUGGCCAUGGUCUUCAUGCCGGCUCAACACUGACAGGUGUAAAUUGCCGAGGCUUAACAAAAG .(((((((((((((....))))))...(((((---((.........)))).(....)..(((.(((.....)))))))))...(((((....)))))....)))))))............ ( -35.20) >consensus ____G________________GAAUCUGCCGG_CGGCAGUCAUAAAGCCCAGGAAACAUGGCCAUGGUCUUCAUGCCGGCUCAACACUGACAGGUGUAAAUUGCCGAGGCUUAACAAAAG .....................((..(((((....)))))))...((((((.(....)..(((.(((.....))))))(((...(((((....))))).....)))).)))))........ (-27.38 = -27.98 + 0.60)


| Location | 11,397,671 – 11,397,791 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.08 |
| Mean single sequence MFE | -36.22 |
| Consensus MFE | -33.32 |
| Energy contribution | -33.32 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.58 |
| SVM RNA-class probability | 0.965644 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11397671 120 + 27905053 CAUAAAGCCCAGGAAACAUGGCCAUGGUCUUCAUACCGGCUCAACACUGACAGGUGUAAAUUGCCGAGGCUUAACAAAAGCCACUGCAGAGCUUCGAGUAAACAGGAGGAGCAGGAUGUG ((((....((((....).)))((.((.(((((.(((((((((.(((((....)))))....(((.(.(((((.....))))).).)))))))..)).))).....))))).)))).)))) ( -36.50) >DroSec_CAF1 145370 120 + 1 CAUAAAGCCCAGGAAACAUGGCCAUGGUCUUCAUGCCGGCUCAACACUGACAGGUGUAAAUUGCCGAGGCUUAACAAAAGCCACUGCAGAGCUUUGAGUAAACAGGAGGAGCAGGAUGUG ((((....((((....).)))((.((.(((((.((...((((((((((....)))))...((((.(.(((((.....))))).).)))).....)))))...)).))))).)))).)))) ( -37.40) >DroSim_CAF1 136452 120 + 1 CAUAAAGCCCAGGAAACAUGGCCAUGGUCUUCAUGCCGGCUCAACACUGACAGGUGUAAAUUGCCGAGGCUUAACAAAAGCCACUGCAGAGCUUUGAGUAAACAGGAGGAGCAGGAUGUG ((((....((((....).)))((.((.(((((.((...((((((((((....)))))...((((.(.(((((.....))))).).)))).....)))))...)).))))).)))).)))) ( -37.40) >DroEre_CAF1 133541 119 + 1 CAUAAAGCCCAGGAAACAUGGCCAUGGUCUUCAUGCCGGCUCAACACUGACAGGUGUAAAUUGCCGAGGCUUAACAAAAGCCACUGGG-AGCUUUGAGUAAACAGGAGGAGCAGGAUGUA ........((((....).)))((.((.(((((.((...((((((..((..((((((.....))))..(((((.....)))))..))..-))..))))))...)).))))).))))..... ( -35.40) >DroYak_CAF1 136345 120 + 1 CAUAAAGCCCAGGAAACAUGGCCAUGGUCUUCAUGCCGGCUCAACACUGACAGGUGUAAAUUGCCGAGGCUUAACAAAAGCCACUGCGAAGCUUUCAGUAAACAGGAGGAGCAGGAUGUA ......(((..(....)..)))((((.....)))).((((...(((((....))))).....)))).(((((.....))))).((((....(((((........))))).))))...... ( -34.40) >consensus CAUAAAGCCCAGGAAACAUGGCCAUGGUCUUCAUGCCGGCUCAACACUGACAGGUGUAAAUUGCCGAGGCUUAACAAAAGCCACUGCAGAGCUUUGAGUAAACAGGAGGAGCAGGAUGUG ........((((....).)))((.((.(((((....((((...(((((....))))).....)))).(((((.....))))).(((....((.....))...)))))))).))))..... (-33.32 = -33.32 + 0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:23:57 2006