
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 11,388,051 – 11,388,243 |
| Length | 192 |
| Max. P | 0.999046 |

| Location | 11,388,051 – 11,388,171 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.83 |
| Mean single sequence MFE | -26.05 |
| Consensus MFE | -23.08 |
| Energy contribution | -23.48 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.807070 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11388051 120 + 27905053 GUUCCACAAACUCUCAACUUUCUUCAUCAGAUAUUUGCCACCUAAUUGUUUGGACAAUUAAAUCAUUAAAAUGUUCAAUGAGGAAACAAGUUCUUUUGUUUCCCGAUUUUGGCGGCCUGC ...((.((((.((..........((((..(((((((......(((((((....)))))))........)))))))..))))(((((((((....))))))))).)).))))..))..... ( -24.54) >DroSec_CAF1 135826 120 + 1 GUUCCAGAAACUUUCAACUUUCUUCAUCAGCGAUGUGCCACCUAAUUGUUUGGACAAUUAAAUCAUUAAAAUGUUCAAUGAGGAAACAAGUUCUUUUGUUUCCCGAUUUUGGCGGCCUGC ....(((.................((((...)))).(((.((.(((((.(((((((.((((....))))..)))))))...(((((((((....))))))))))))))..)).)))))). ( -26.60) >DroSim_CAF1 126925 120 + 1 GUUCCAGAAACUUUCAACUUUCUUCAUCAGCGAUUUGCCACCUAAUUGCUUGGACAAUUAAAUCAUUAAAAUGUUCAAUGAGGAAACAAGUUCUUUUGUUUCCCGAUUUUGGCGGCCUGC (((((((((..............((((.(((((((........)))))))((((((.((((....))))..))))))))))(((((((((....)))))))))...)))))).))).... ( -25.80) >DroEre_CAF1 124310 119 + 1 GCUCCAGAAACUCUCAACUUUCUUCAUCAGCGAUUUGCCACCUAAUUGUUUGGACAAUUAAAUCAUUAAAAUGUUCAAUGAGGA-ACAAGUUCUUUUGUUUCCCGAUUUUGGCGGCCCGC .....(((((........)))))......(((..((((((...(((((.(((((((.((((....))))..)))))))...(((-(((((....)))))))).))))).))))))..))) ( -23.90) >DroYak_CAF1 126560 120 + 1 GUGCCAGAGACUCUCAACUUUCUUCAUCAGCGAUUUGCCACCUAAUUGUUUGGACAAUUAAAUCAUUAAAAUGUUCAAUGAGGAAACAAGUUCUUUUGUUUCCCGACUUUGGCGGCCUGC .((((((((..............((((.(((((((........)))))))((((((.((((....))))..))))))))))(((((((((....)))))))))...))))))))...... ( -29.40) >consensus GUUCCAGAAACUCUCAACUUUCUUCAUCAGCGAUUUGCCACCUAAUUGUUUGGACAAUUAAAUCAUUAAAAUGUUCAAUGAGGAAACAAGUUCUUUUGUUUCCCGAUUUUGGCGGCCUGC ....................................(((.((.(((((.(((((((.((((....))))..)))))))...(((((((((....))))))))))))))..)).))).... (-23.08 = -23.48 + 0.40)



| Location | 11,388,051 – 11,388,171 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.83 |
| Mean single sequence MFE | -31.28 |
| Consensus MFE | -29.16 |
| Energy contribution | -29.24 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.93 |
| SVM decision value | 3.34 |
| SVM RNA-class probability | 0.999046 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11388051 120 - 27905053 GCAGGCCGCCAAAAUCGGGAAACAAAAGAACUUGUUUCCUCAUUGAACAUUUUAAUGAUUUAAUUGUCCAAACAAUUAGGUGGCAAAUAUCUGAUGAAGAAAGUUGAGAGUUUGUGGAAC ((((((((((.......((((((((......))))))))(((((((.....)))))))..(((((((....))))))))))))).....(((.....)))...........))))..... ( -31.00) >DroSec_CAF1 135826 120 - 1 GCAGGCCGCCAAAAUCGGGAAACAAAAGAACUUGUUUCCUCAUUGAACAUUUUAAUGAUUUAAUUGUCCAAACAAUUAGGUGGCACAUCGCUGAUGAAGAAAGUUGAAAGUUUCUGGAAC .(((((((((.......((((((((......))))))))(((((((.....)))))))..(((((((....))))))))))))).((((...)))).................))).... ( -32.80) >DroSim_CAF1 126925 120 - 1 GCAGGCCGCCAAAAUCGGGAAACAAAAGAACUUGUUUCCUCAUUGAACAUUUUAAUGAUUUAAUUGUCCAAGCAAUUAGGUGGCAAAUCGCUGAUGAAGAAAGUUGAAAGUUUCUGGAAC .(((((((((.......((((((((......))))))))(((((((.....)))))))..(((((((....)))))))))))))...(((((.........)).)))......))).... ( -31.80) >DroEre_CAF1 124310 119 - 1 GCGGGCCGCCAAAAUCGGGAAACAAAAGAACUUGU-UCCUCAUUGAACAUUUUAAUGAUUUAAUUGUCCAAACAAUUAGGUGGCAAAUCGCUGAUGAAGAAAGUUGAGAGUUUCUGGAGC ((((((((((.......((((.(((......))))-)))(((((((.....)))))))..(((((((....)))))))))))))...))))......(((((.(....).)))))..... ( -29.30) >DroYak_CAF1 126560 120 - 1 GCAGGCCGCCAAAGUCGGGAAACAAAAGAACUUGUUUCCUCAUUGAACAUUUUAAUGAUUUAAUUGUCCAAACAAUUAGGUGGCAAAUCGCUGAUGAAGAAAGUUGAGAGUCUCUGGCAC .(((((((((.......((((((((......))))))))(((((((.....)))))))..(((((((....)))))))))))))...(((((.........)).)))......))).... ( -31.50) >consensus GCAGGCCGCCAAAAUCGGGAAACAAAAGAACUUGUUUCCUCAUUGAACAUUUUAAUGAUUUAAUUGUCCAAACAAUUAGGUGGCAAAUCGCUGAUGAAGAAAGUUGAGAGUUUCUGGAAC .(((((((((.......((((((((......))))))))(((((((.....)))))))..(((((((....)))))))))))))......((.....))..............))).... (-29.16 = -29.24 + 0.08)

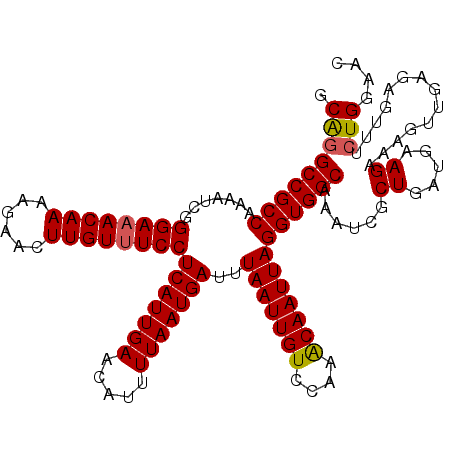

| Location | 11,388,091 – 11,388,210 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.64 |
| Mean single sequence MFE | -26.71 |
| Consensus MFE | -23.14 |
| Energy contribution | -23.22 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.702668 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11388091 119 - 27905053 -CCCAGCCCAAAGUCGCCAGCCAAAUAAUUGAUGAAAGUGGCAGGCCGCCAAAAUCGGGAAACAAAAGAACUUGUUUCCUCAUUGAACAUUUUAAUGAUUUAAUUGUCCAAACAAUUAGG -...........((.(((.((((.....((.....)).)))).))).))........((((((((......))))))))(((((((.....))))))).((((((((....)))))))). ( -27.20) >DroSec_CAF1 135866 119 - 1 -CCCAGCCCAAAGUCGCCGGCUAAAUAAUUGAUGAAAGUGGCAGGCCGCCAAAAUCGGGAAACAAAAGAACUUGUUUCCUCAUUGAACAUUUUAAUGAUUUAAUUGUCCAAACAAUUAGG -...((((..........)))).......((((....((((....))))....))))((((((((......))))))))(((((((.....))))))).((((((((....)))))))). ( -28.10) >DroSim_CAF1 126965 119 - 1 -CCCAGCCCAAAGUCGCCGGCUAAAUAAUUGAUGAAAGUGGCAGGCCGCCAAAAUCGGGAAACAAAAGAACUUGUUUCCUCAUUGAACAUUUUAAUGAUUUAAUUGUCCAAGCAAUUAGG -...((((..........)))).......((((....((((....))))....))))((((((((......))))))))(((((((.....))))))).((((((((....)))))))). ( -28.40) >DroEre_CAF1 124350 117 - 1 -UCUAGCCCAAAGUCCGCAGC-AAAUAAUUGAUGAAAGUAGCGGGCCGCCAAAAUCGGGAAACAAAAGAACUUGU-UCCUCAUUGAACAUUUUAAUGAUUUAAUUGUCCAAACAAUUAGG -...........((((((.((-...............)).))))))...........((((.(((......))))-)))(((((((.....))))))).((((((((....)))))))). ( -22.26) >DroYak_CAF1 126600 119 - 1 CUCUGGCCCAAAGUCCCCGGC-AAAUAAUUGAUGAAAGUGGCAGGCCGCCAAAGUCGGGAAACAAAAGAACUUGUUUCCUCAUUGAACAUUUUAAUGAUUUAAUUGUCCAAACAAUUAGG .....(((..........)))-..........(((...((((.....))))...)))((((((((......))))))))(((((((.....))))))).((((((((....)))))))). ( -27.60) >consensus _CCCAGCCCAAAGUCGCCGGC_AAAUAAUUGAUGAAAGUGGCAGGCCGCCAAAAUCGGGAAACAAAAGAACUUGUUUCCUCAUUGAACAUUUUAAUGAUUUAAUUGUCCAAACAAUUAGG .............................((((....((((....))))....))))((((((((......))))))))(((((((.....))))))).((((((((....)))))))). (-23.14 = -23.22 + 0.08)



| Location | 11,388,131 – 11,388,243 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.01 |
| Mean single sequence MFE | -33.91 |
| Consensus MFE | -29.12 |
| Energy contribution | -29.16 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.599497 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11388131 112 + 27905053 AGGAAACAAGUUCUUUUGUUUCCCGAUUUUGGCGGCCUGCCACUUUCAUCAAUUAUUUGGCUGGCGACUUUGGGCUGGG--------GCUUUGCGGGAAAUUCCUUGUUUGUCCGCAAAC .(((((((((....)))))))))(((..(((.(((((.....................))))).)))..)))(((....--------)))(((((((((((.....)))).))))))).. ( -34.10) >DroSec_CAF1 135906 112 + 1 AGGAAACAAGUUCUUUUGUUUCCCGAUUUUGGCGGCCUGCCACUUUCAUCAAUUAUUUAGCCGGCGACUUUGGGCUGGG--------GCUUUGCGGGAAAUUCCUUGUUUGUCCGCAAAC .(((((((((....))))))))).((...((((.....))))...)).............(((((........))))).--------..((((((((((((.....)))).)))))))). ( -34.00) >DroSim_CAF1 127005 112 + 1 AGGAAACAAGUUCUUUUGUUUCCCGAUUUUGGCGGCCUGCCACUUUCAUCAAUUAUUUAGCCGGCGACUUUGGGCUGGG--------GCUUUGCGGGAAAUUCCUUGUUUGUCCGCAAAC .(((((((((....))))))))).((...((((.....))))...)).............(((((........))))).--------..((((((((((((.....)))).)))))))). ( -34.00) >DroEre_CAF1 124390 110 + 1 AGGA-ACAAGUUCUUUUGUUUCCCGAUUUUGGCGGCCCGCUACUUUCAUCAAUUAUUU-GCUGCGGACUUUGGGCUAGA--------GCUUUGCGGCAAAUUCCUUGUUUGUCCGCAAAC .(((-(((((....))))))))((((..((.(((((......................-))))).))..))))((....--------))((((((((((((.....))))).))))))). ( -32.15) >DroYak_CAF1 126640 119 + 1 AGGAAACAAGUUCUUUUGUUUCCCGACUUUGGCGGCCUGCCACUUUCAUCAAUUAUUU-GCCGGGGACUUUGGGCCAGAGAAAUGGGGCUUUGCGGGAAAUUCCUUGUUUGUCCGCAAAC .(((((((((....)))))))))...(((((((((....)).................-.(((....)...)))))))))...((((((...(((((......)))))..)))).))... ( -35.30) >consensus AGGAAACAAGUUCUUUUGUUUCCCGAUUUUGGCGGCCUGCCACUUUCAUCAAUUAUUU_GCCGGCGACUUUGGGCUGGG________GCUUUGCGGGAAAUUCCUUGUUUGUCCGCAAAC .(((((((((....))))))))).((...((((.....))))...))........(((((((.(......).)))))))..........((((((((((((.....)))).)))))))). (-29.12 = -29.16 + 0.04)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:23:39 2006