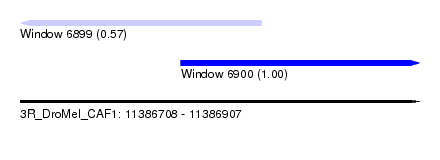
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 11,386,708 – 11,386,907 |
| Length | 199 |
| Max. P | 0.999438 |

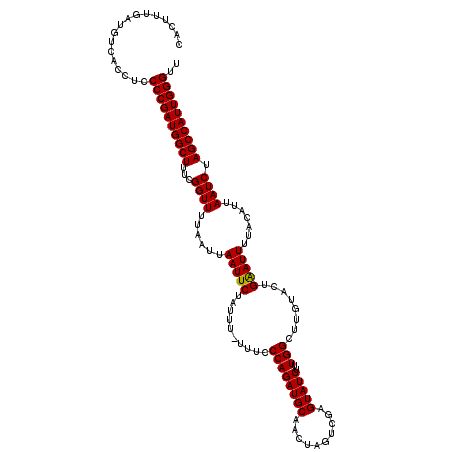
| Location | 11,386,708 – 11,386,828 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.33 |
| Mean single sequence MFE | -29.33 |
| Consensus MFE | -25.12 |
| Energy contribution | -25.28 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.29 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.566674 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11386708 120 - 27905053 AAUUAAAACCGAAAGCCAUCGGGGAGGUGACAUCAAAGUGUUUCUUCGUUCUGGCUUUGGAAUCUCGGGUCCUCCUGAGCAAAUACCAAAAAACUUCAAUUAAUGUAAGCAAAUGAAGCU ........((.(((((((..((.((((.(((((....))))).)))).)).)))))))))...((((((....))))))..............(((((.....((....))..))))).. ( -29.30) >DroSec_CAF1 134456 120 - 1 AAUUAAAACCGAAAGCCAUCGGGGAGGUGACAUCAAAGUGUUUCUUUGUUCUGGCUUUGGAUUCUCGGGUCCUCCAGAGCAAAUACCAAAGAACUUCAAUUAAUGUAAGCAAAUGAAGCU .(((((..((((......)))).((((((((((....)))))..((((((((((....(((((....))))).)))))))))).........)))))..)))))...(((.......))) ( -30.90) >DroSim_CAF1 125577 120 - 1 AAUUAAAACCGAAAGCCAUCGGGGAGGUGACAUCAAAGUGUUUCUUUGUUCUGGCUUUGGAUUCUCGGGUCCUCCUGAGCAAAUACCAAAGAACUUCAAUUAAUGUAAGCAAAUGAAGCU .(((((..((((......)))).(((((..((.(((((.....)))))...)).(((((((((((((((....))))))..))).)))))).)))))..)))))...(((.......))) ( -29.50) >DroEre_CAF1 122989 120 - 1 AAUUAAAACCGAAAGCCAUCGGGGAAGUGACAUCAAAGUGUUUCUUUAUCCUGGCUUGGGAUUCCCGGGUCCUCCUGAGCAAAUACCAAAAAACUUCAAUUAAUGCAAGCAAAUGAAGCU .(((((..((((......)))).(((((.........(((((((((......((((((((...)))))))).....))).))))))......)))))..)))))...(((.......))) ( -25.86) >DroYak_CAF1 125183 120 - 1 AAUUAAAACCGAAAGCCAUCGGGGAAGUGACAUCAAAGUGUUCCUUUGUCCUGGCUUUGGAUGCCCGGGUCCUCCUGAGCAAAUACCAAAAAACUUCAAUUAAUGUAAGCAAAUGAAGCU ........((.(((((((..((.((((..((((....))))..)))).)).))))))))).(((.((((....)))).)))............(((((.....((....))..))))).. ( -31.10) >consensus AAUUAAAACCGAAAGCCAUCGGGGAGGUGACAUCAAAGUGUUUCUUUGUUCUGGCUUUGGAUUCUCGGGUCCUCCUGAGCAAAUACCAAAAAACUUCAAUUAAUGUAAGCAAAUGAAGCU ........((.(((((((..((.((((.(((((....))))).)))).)).)))))))))...((((((....))))))..............(((((.....((....))..))))).. (-25.12 = -25.28 + 0.16)

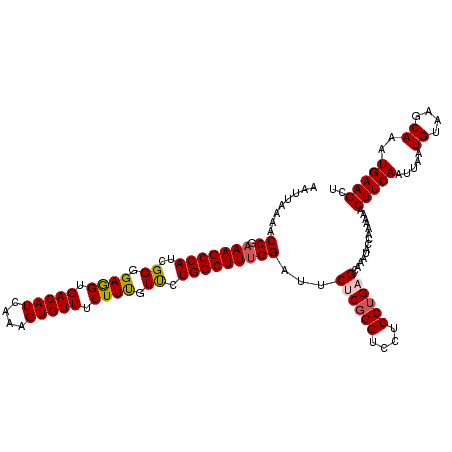
| Location | 11,386,788 – 11,386,907 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.16 |
| Mean single sequence MFE | -28.39 |
| Consensus MFE | -25.87 |
| Energy contribution | -25.63 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.91 |
| SVM decision value | 3.60 |
| SVM RNA-class probability | 0.999438 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11386788 119 + 27905053 CACUUUGAUGUCACCUCCCCGAUGGCUUUCGGUUUUAAUUAAUUCUAUUU-UUUCCCAGAUGCAACUAGUCGAGUAUCUUUGGCUUGUACUGAAUUUUACAUUAAUCUAGCCAUUGGGUU .................((((((((((...((((......(((((.....-...((.((((((..........))))))..))........))))).......)))).)))))))))).. ( -27.41) >DroSec_CAF1 134536 120 + 1 CACUUUGAUGUCACCUCCCCGAUGGCUUUCGGUUUUAAUUAAUUCUAUUUUUUUCCCAGAUGCAACUAGUCGAGUAUCUUUGGCUUGUAGUGAAUUUUACAUUAAUCUAGCCAUUGGGUU .................((((((((((...((((......(((((.(((.....((.((((((..........))))))..)).....)))))))).......)))).)))))))))).. ( -27.52) >DroSim_CAF1 125657 120 + 1 CACUUUGAUGUCACCUCCCCGAUGGCUUUCGGUUUUAAUUAAUUCUAUUUUUUUCCCAGAUGCAACUAGUCGGGUAUCUUUGGUUUGUACUGAAUUUUACAUUAAUCUAGCCAUUGGGUU .................((((((((((...((((......(((((.........((.((((((..........))))))..))........))))).......)))).)))))))))).. ( -27.45) >DroEre_CAF1 123069 119 + 1 CACUUUGAUGUCACUUCCCCGAUGGCUUUCGGUUUUAAUUAAUUCUAUUU-UUUCCCAGAUGCAACUAGUCGAGUAUCCUUGGCUUGUACCGGAUUUUACAUUAAUCUAGCCAUUGGGUU .................((((((((((..((((............(((((-......)))))..((.(((((((....))))))).))))))((((.......)))).)))))))))).. ( -29.10) >DroYak_CAF1 125263 119 + 1 CACUUUGAUGUCACUUCCCCGAUGGCUUUCGGUUUUAAUUAAUUCUAUUU-UCGCCCAGAUGCAACUAGUCGAGUAUCUUUGGCUUGUACUGGAUUUUACAUUAAUCUAGCCAUUGGGUU .................((((((((((...((((......(((((.....-..(((.((((((..........))))))..))).......))))).......)))).)))))))))).. ( -30.46) >consensus CACUUUGAUGUCACCUCCCCGAUGGCUUUCGGUUUUAAUUAAUUCUAUUU_UUUCCCAGAUGCAACUAGUCGAGUAUCUUUGGCUUGUACUGAAUUUUACAUUAAUCUAGCCAUUGGGUU .................((((((((((...((((......(((((..........((((((((..........)))))..)))........))))).......)))).)))))))))).. (-25.87 = -25.63 + -0.24)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:23:33 2006