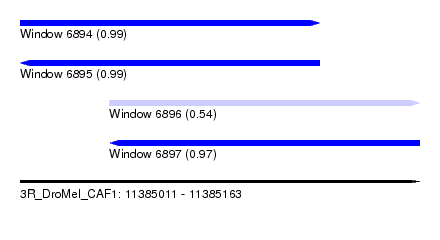
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 11,385,011 – 11,385,163 |
| Length | 152 |
| Max. P | 0.990715 |

| Location | 11,385,011 – 11,385,125 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.91 |
| Mean single sequence MFE | -32.68 |
| Consensus MFE | -31.28 |
| Energy contribution | -31.32 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.96 |
| SVM decision value | 2.23 |
| SVM RNA-class probability | 0.990715 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
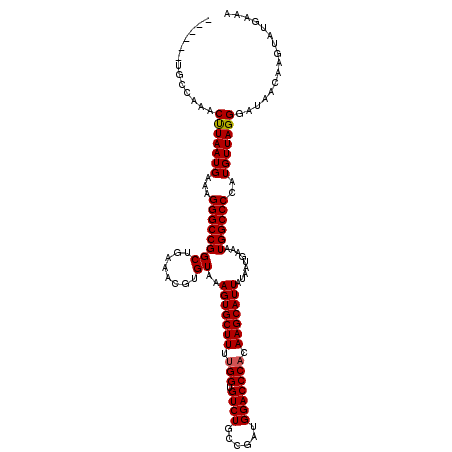
>3R_DroMel_CAF1 11385011 114 + 27905053 ------UGCCAAACUUAAUGAAAGGGCCGGCUGAAACGUGUAAAGUGCUUUUGGUGUCUGCCGAUGGACCCACAAGCAUUAUAAUGAAAUGGCCCCAUGUUAGGGAUAACAAGUAUGAAA ------...((.((((.......((((((((........))..(((((((.(((.((((......))))))).))))))).........))))))...((((....)))))))).))... ( -31.90) >DroSec_CAF1 132720 114 + 1 ------UGCCAAACUUAAUGAAAGGGCCGGCUGAAACGUGUAAAGUGCUUUUGGUGUCUGCCGAUGGACCCACAAGCAUUAUAAUGAAAUGGCCCCAUGUUAGGGAUAACAAGUAUGAAA ------...((.((((.......((((((((........))..(((((((.(((.((((......))))))).))))))).........))))))...((((....)))))))).))... ( -31.90) >DroSim_CAF1 123842 114 + 1 ------UGCCAAACUUAAUGAAAGGGCCGGCUGAAACGUGUAAAGUGCUUUUGGUGUCUGCCGAUGGACCCACAAGCAUUAUAAUGAAAUGGCCCCAUGUUAGGGAUAACAAGUAUGAAA ------...((.((((.......((((((((........))..(((((((.(((.((((......))))))).))))))).........))))))...((((....)))))))).))... ( -31.90) >DroEre_CAF1 120154 114 + 1 ------UGCCAAACCUAAUGAAAGGGCCGGCUGAAACGUGUAAAGUGCUUUCGGUGUCUGCCGAUGGACCCACAAGCAUUAUAAUGAAAUGGCCCCAUGUUAGGGAUAACAAGUAUGAAA ------.......(((((((...((((((((........))..(((((((..((.((((......))))))..))))))).........))))))..)))))))................ ( -31.10) >DroYak_CAF1 123436 120 + 1 UGCCUAUGCCAAACUUAAUGAAGGGGCCGGCUGAAACGUGUAAAGUGCUUUUGGUGUCUGCCGAUGGACCCACAAGCAUUAUAAUGAAAUGGCCCCAUGUUAGGGAUAACAAGUAUGAAA ....(((((.............(((((((((........))..(((((((.(((.((((......))))))).))))))).........))))))).(((((....))))).)))))... ( -36.60) >consensus ______UGCCAAACUUAAUGAAAGGGCCGGCUGAAACGUGUAAAGUGCUUUUGGUGUCUGCCGAUGGACCCACAAGCAUUAUAAUGAAAUGGCCCCAUGUUAGGGAUAACAAGUAUGAAA .............(((((((...((((((((........))..(((((((.(((.((((......))))))).))))))).........))))))..)))))))................ (-31.28 = -31.32 + 0.04)

| Location | 11,385,011 – 11,385,125 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.91 |
| Mean single sequence MFE | -29.06 |
| Consensus MFE | -28.10 |
| Energy contribution | -27.62 |
| Covariance contribution | -0.48 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.97 |
| SVM decision value | 2.18 |
| SVM RNA-class probability | 0.989701 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11385011 114 - 27905053 UUUCAUACUUGUUAUCCCUAACAUGGGGCCAUUUCAUUAUAAUGCUUGUGGGUCCAUCGGCAGACACCAAAAGCACUUUACACGUUUCAGCCGGCCCUUUCAUUAAGUUUGGCA------ ..(((.((((((((....))))..((((((............(((((.((((((........))).))).)))))........((....)).))))))......)))).)))..------ ( -27.30) >DroSec_CAF1 132720 114 - 1 UUUCAUACUUGUUAUCCCUAACAUGGGGCCAUUUCAUUAUAAUGCUUGUGGGUCCAUCGGCAGACACCAAAAGCACUUUACACGUUUCAGCCGGCCCUUUCAUUAAGUUUGGCA------ ..(((.((((((((....))))..((((((............(((((.((((((........))).))).)))))........((....)).))))))......)))).)))..------ ( -27.30) >DroSim_CAF1 123842 114 - 1 UUUCAUACUUGUUAUCCCUAACAUGGGGCCAUUUCAUUAUAAUGCUUGUGGGUCCAUCGGCAGACACCAAAAGCACUUUACACGUUUCAGCCGGCCCUUUCAUUAAGUUUGGCA------ ..(((.((((((((....))))..((((((............(((((.((((((........))).))).)))))........((....)).))))))......)))).)))..------ ( -27.30) >DroEre_CAF1 120154 114 - 1 UUUCAUACUUGUUAUCCCUAACAUGGGGCCAUUUCAUUAUAAUGCUUGUGGGUCCAUCGGCAGACACCGAAAGCACUUUACACGUUUCAGCCGGCCCUUUCAUUAGGUUUGGCA------ .........(((((..(((((...((((((............(((((.((((((........))).))).)))))........((....)).))))))....)))))..)))))------ ( -32.90) >DroYak_CAF1 123436 120 - 1 UUUCAUACUUGUUAUCCCUAACAUGGGGCCAUUUCAUUAUAAUGCUUGUGGGUCCAUCGGCAGACACCAAAAGCACUUUACACGUUUCAGCCGGCCCCUUCAUUAAGUUUGGCAUAGGCA ...((.((((((((....))))..((((((............(((((.((((((........))).))).)))))........((....)).))))))......)))).))((....)). ( -30.50) >consensus UUUCAUACUUGUUAUCCCUAACAUGGGGCCAUUUCAUUAUAAUGCUUGUGGGUCCAUCGGCAGACACCAAAAGCACUUUACACGUUUCAGCCGGCCCUUUCAUUAAGUUUGGCA______ ..(((.((((((((....))))..((((((............(((((.((((((........))).))).)))))........((....)).))))))......)))).)))........ (-28.10 = -27.62 + -0.48)

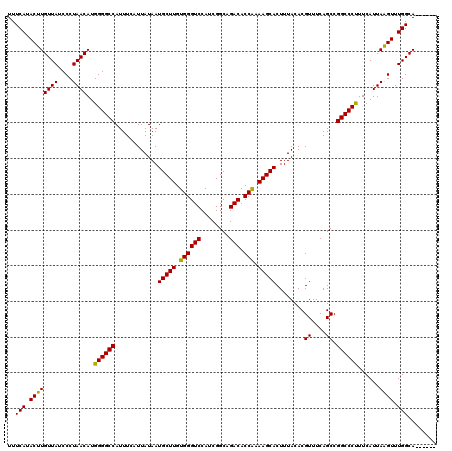
| Location | 11,385,045 – 11,385,163 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.31 |
| Mean single sequence MFE | -35.12 |
| Consensus MFE | -32.34 |
| Energy contribution | -33.14 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.13 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.537047 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
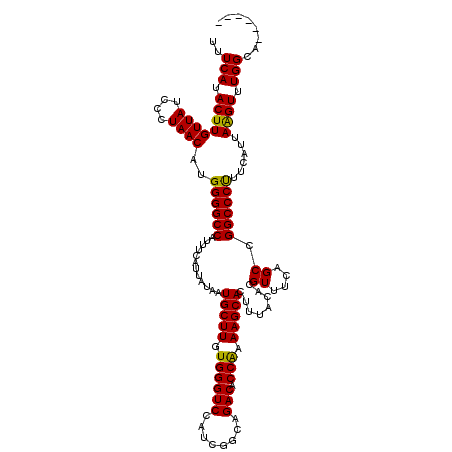
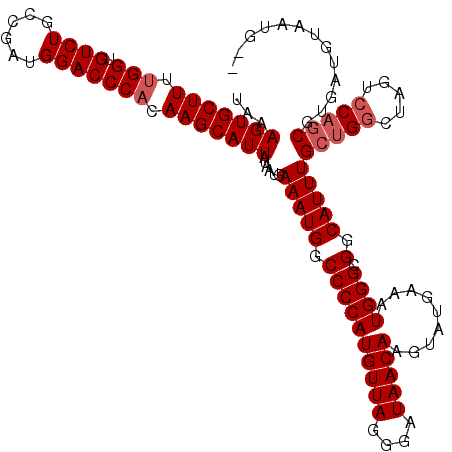
>3R_DroMel_CAF1 11385045 118 + 27905053 UAAAGUGCUUUUGGUGUCUGCCGAUGGACCCACAAGCAUUAUAAUGAAAUGGCCCCAUGUUAGGGAUAACAAGUAUGAAAUGGGCGGCAUUUGCCGGCUAGUCCAUCGUGAUGUAAUG-- ...(((((((.(((.((((......))))))).)))))))...((((..(((((((......)))...............(((.((((....)))).)))).))))))).........-- ( -34.70) >DroSec_CAF1 132754 120 + 1 UAAAGUGCUUUUGGUGUCUGCCGAUGGACCCACAAGCAUUAUAAUGAAAUGGCCCCAUGUUAGGGAUAACAAGUAUGAAAUGGGCGGCAUUUGCUGGCUAGUCCAGCGUGAUGUAAUAGC ...(((((((.(((.((((......))))))).)))))))......(((((.((((((((((....))))).........)))).).)))))(((((.....)))))............. ( -35.80) >DroSim_CAF1 123876 120 + 1 UAAAGUGCUUUUGGUGUCUGCCGAUGGACCCACAAGCAUUAUAAUGAAAUGGCCCCAUGUUAGGGAUAACAAGUAUGAAAUGGGCGGCAUUUGCUGGCUAGUCCAGCGUGAUGUAAUAGC ...(((((((.(((.((((......))))))).)))))))......(((((.((((((((((....))))).........)))).).)))))(((((.....)))))............. ( -35.80) >DroEre_CAF1 120188 118 + 1 UAAAGUGCUUUCGGUGUCUGCCGAUGGACCCACAAGCAUUAUAAUGAAAUGGCCCCAUGUUAGGGAUAACAAGUAUGAAAUGGGCGGCAUUUGCUGGCUAGUACAGCGUGAUGUGAUG-- ....(((((.(((((....))))).))...)))..(((((((....(((((.((((((((((....))))).........)))).).)))))((((.......)))))))))))....-- ( -32.80) >DroYak_CAF1 123476 118 + 1 UAAAGUGCUUUUGGUGUCUGCCGAUGGACCCACAAGCAUUAUAAUGAAAUGGCCCCAUGUUAGGGAUAACAAGUAUGAAAUGGGCGGCAUUUGCUGGCUAGCCCAGCGUGAUGUGAUG-- ...(((((((.(((.((((......))))))).)))))))........((.((..((((((.(((....((....))...(((.((((....)))).))).)))))))))..)).)).-- ( -36.50) >consensus UAAAGUGCUUUUGGUGUCUGCCGAUGGACCCACAAGCAUUAUAAUGAAAUGGCCCCAUGUUAGGGAUAACAAGUAUGAAAUGGGCGGCAUUUGCUGGCUAGUCCAGCGUGAUGUAAUG__ ...(((((((.(((.((((......))))))).)))))))......(((((.((((((((((....))))).........)))).).)))))(((((.....)))))............. (-32.34 = -33.14 + 0.80)

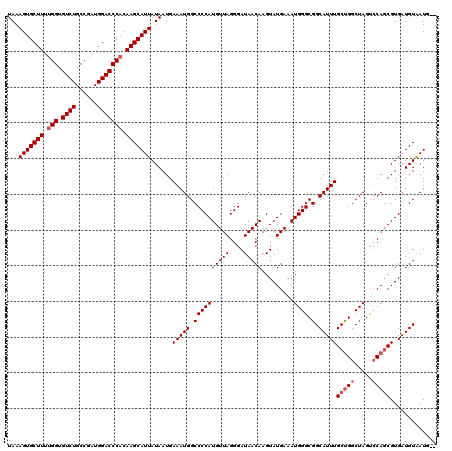
| Location | 11,385,045 – 11,385,163 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.31 |
| Mean single sequence MFE | -29.42 |
| Consensus MFE | -26.94 |
| Energy contribution | -27.02 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.72 |
| SVM RNA-class probability | 0.974078 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
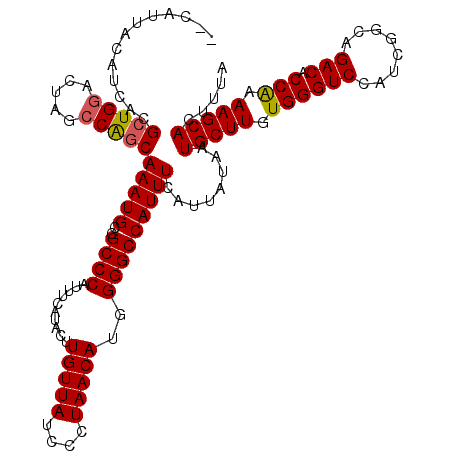
>3R_DroMel_CAF1 11385045 118 - 27905053 --CAUUACAUCACGAUGGACUAGCCGGCAAAUGCCGCCCAUUUCAUACUUGUUAUCCCUAACAUGGGGCCAUUUCAUUAUAAUGCUUGUGGGUCCAUCGGCAGACACCAAAAGCACUUUA --..........((((((((((((.(((....)))((((..........(((((....)))))..))))..............)))....)))))))))..................... ( -29.50) >DroSec_CAF1 132754 120 - 1 GCUAUUACAUCACGCUGGACUAGCCAGCAAAUGCCGCCCAUUUCAUACUUGUUAUCCCUAACAUGGGGCCAUUUCAUUAUAAUGCUUGUGGGUCCAUCGGCAGACACCAAAAGCACUUUA (((..........(((((.....)))))...(((((.............(((((....)))))((((.((((..(((....)))...)))).)))).))))).........)))...... ( -31.60) >DroSim_CAF1 123876 120 - 1 GCUAUUACAUCACGCUGGACUAGCCAGCAAAUGCCGCCCAUUUCAUACUUGUUAUCCCUAACAUGGGGCCAUUUCAUUAUAAUGCUUGUGGGUCCAUCGGCAGACACCAAAAGCACUUUA (((..........(((((.....)))))...(((((.............(((((....)))))((((.((((..(((....)))...)))).)))).))))).........)))...... ( -31.60) >DroEre_CAF1 120188 118 - 1 --CAUCACAUCACGCUGUACUAGCCAGCAAAUGCCGCCCAUUUCAUACUUGUUAUCCCUAACAUGGGGCCAUUUCAUUAUAAUGCUUGUGGGUCCAUCGGCAGACACCGAAAGCACUUUA --.......((..((((.......))))...(((((.............(((((....)))))((((.((((..(((....)))...)))).)))).)))))......)).......... ( -25.00) >DroYak_CAF1 123476 118 - 1 --CAUCACAUCACGCUGGGCUAGCCAGCAAAUGCCGCCCAUUUCAUACUUGUUAUCCCUAACAUGGGGCCAUUUCAUUAUAAUGCUUGUGGGUCCAUCGGCAGACACCAAAAGCACUUUA --...........(((((.....)))))(((((..((((..........(((((....)))))..)))))))))........(((((.((((((........))).))).)))))..... ( -29.40) >consensus __CAUUACAUCACGCUGGACUAGCCAGCAAAUGCCGCCCAUUUCAUACUUGUUAUCCCUAACAUGGGGCCAUUUCAUUAUAAUGCUUGUGGGUCCAUCGGCAGACACCAAAAGCACUUUA .............(((((.....)))))(((((..((((..........(((((....)))))..)))))))))........(((((.((((((........))).))).)))))..... (-26.94 = -27.02 + 0.08)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:23:29 2006