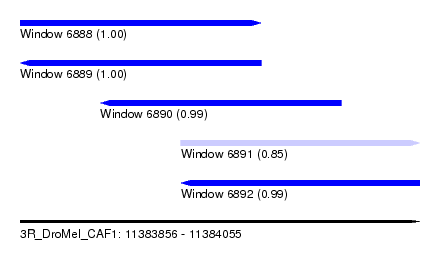
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 11,383,856 – 11,384,055 |
| Length | 199 |
| Max. P | 0.999736 |

| Location | 11,383,856 – 11,383,976 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.83 |
| Mean single sequence MFE | -32.30 |
| Consensus MFE | -29.58 |
| Energy contribution | -29.66 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.92 |
| SVM decision value | 3.32 |
| SVM RNA-class probability | 0.999007 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11383856 120 + 27905053 AGUUUUUACACUUUCCAGCAGCGUUUUUGGAGUUUCAGCGUUUCCUCAACGUUUCCGAAACUACUUACGAUGCCUUGAACUUCAUUUCCCGGCCAGGACCUUUUUUGGCCUUUGGCCAUA ............(((..(((.(((....((((((((((((((.....))))))...)))))).)).))).)))...)))...........(((((((.((......))))..)))))... ( -29.10) >DroSec_CAF1 131583 120 + 1 AGUUUUUACACUUUCCAGCAGCGUUUUUGGAGUUUCGGCGUUUCCUCAUCGUUUCCGAAACUACUUACGAUGCCUUGAACUUCAUUUCCCGGCCAGGACCUUUUCUGGCCUUUGGCCAUA ..............((((((.(((....((((((((((((.........))...)))))))).)).))).))).................((((((((....))))))))..)))..... ( -30.80) >DroSim_CAF1 122684 120 + 1 AGUUUUUACACUUUCCAGCAGCGUUUUUGGAGUUUCGGCGUUUCCUCAACGUUUCCGAAACUACUUACGAUGCCUUGAACUUCAUUUCCCGGCCAGGACCUUUUCUGGCCUUUGGCCAUA ..............((((((.(((....((((((((((((((.....))))...)))))))).)).))).))).................((((((((....))))))))..)))..... ( -33.30) >DroEre_CAF1 119055 120 + 1 AGUUUUUACAGUUUCUAACAGCGUUUCUGGAGUUUCGGCGUUUCCUCAACGUUUCCGAAACUACUUACGAUGCCUUGAACUUCAUUUCCCGGCCAGGACCUUUUCUGGCCUCUGGCCAUA .........(((((......(((((...((((((((((((((.....))))...)))))))).))...)))))...))))).........(((((((.((......)).).))))))... ( -33.50) >DroYak_CAF1 121802 120 + 1 AGUUUGUACAGUUUCCAGCAGCGUUUUUGGAGUUUCGGCGUUUCCUCAACGUUUCCGAAACUACUUACGAUGCCUUGAACUUCAUUUCCCGGCCAGGACCUUUUCUGGCCUUUGGCCAUA ....((..((((((...(((.(((....((((((((((((((.....))))...)))))))).)).))).)))...))))..........((((((((....))))))))..))..)).. ( -34.80) >consensus AGUUUUUACACUUUCCAGCAGCGUUUUUGGAGUUUCGGCGUUUCCUCAACGUUUCCGAAACUACUUACGAUGCCUUGAACUUCAUUUCCCGGCCAGGACCUUUUCUGGCCUUUGGCCAUA ..............(((...(((((...((((((((((((((.....))))...)))))))).))...))))).................((((((((....))))))))..)))..... (-29.58 = -29.66 + 0.08)

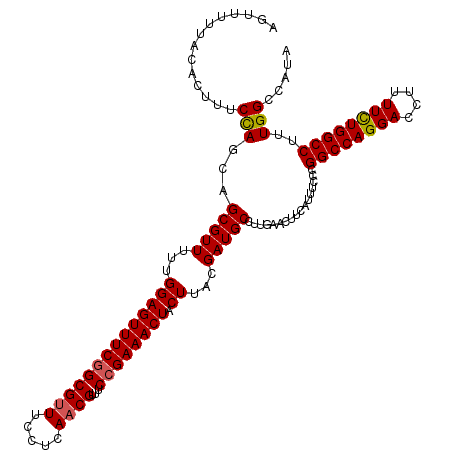
| Location | 11,383,856 – 11,383,976 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.83 |
| Mean single sequence MFE | -35.64 |
| Consensus MFE | -33.00 |
| Energy contribution | -33.08 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.64 |
| Structure conservation index | 0.93 |
| SVM decision value | 3.97 |
| SVM RNA-class probability | 0.999736 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11383856 120 - 27905053 UAUGGCCAAAGGCCAAAAAAGGUCCUGGCCGGGAAAUGAAGUUCAAGGCAUCGUAAGUAGUUUCGGAAACGUUGAGGAAACGCUGAAACUCCAAAAACGCUGCUGGAAAGUGUAAAAACU ..((((((..((((......)))).))))))(((.((((.((.....)).)))).....(((((((...((((.....))))))))))))))....(((((.......)))))....... ( -33.30) >DroSec_CAF1 131583 120 - 1 UAUGGCCAAAGGCCAGAAAAGGUCCUGGCCGGGAAAUGAAGUUCAAGGCAUCGUAAGUAGUUUCGGAAACGAUGAGGAAACGCCGAAACUCCAAAAACGCUGCUGGAAAGUGUAAAAACU ..((((((..((((......)))).)))))).....((.((((...(((.............(((....)))...(....))))..)))).))...(((((.......)))))....... ( -34.20) >DroSim_CAF1 122684 120 - 1 UAUGGCCAAAGGCCAGAAAAGGUCCUGGCCGGGAAAUGAAGUUCAAGGCAUCGUAAGUAGUUUCGGAAACGUUGAGGAAACGCCGAAACUCCAAAAACGCUGCUGGAAAGUGUAAAAACU (((..((...((((((........)))))).))..)))...(((..((((.(((..(.((((((((...((((.....)))))))))))).)....))).)))).)))(((......))) ( -35.60) >DroEre_CAF1 119055 120 - 1 UAUGGCCAGAGGCCAGAAAAGGUCCUGGCCGGGAAAUGAAGUUCAAGGCAUCGUAAGUAGUUUCGGAAACGUUGAGGAAACGCCGAAACUCCAGAAACGCUGUUAGAAACUGUAAAAACU ..(((((((.((((......)))))))))))........((((...((((.(((..(.((((((((...((((.....)))))))))))).)....))).))))...))))......... ( -37.50) >DroYak_CAF1 121802 120 - 1 UAUGGCCAAAGGCCAGAAAAGGUCCUGGCCGGGAAAUGAAGUUCAAGGCAUCGUAAGUAGUUUCGGAAACGUUGAGGAAACGCCGAAACUCCAAAAACGCUGCUGGAAACUGUACAAACU (((..((...((((((........)))))).))..))).((((...((((.(((..(.((((((((...((((.....)))))))))))).)....))).))))...))))......... ( -37.60) >consensus UAUGGCCAAAGGCCAGAAAAGGUCCUGGCCGGGAAAUGAAGUUCAAGGCAUCGUAAGUAGUUUCGGAAACGUUGAGGAAACGCCGAAACUCCAAAAACGCUGCUGGAAAGUGUAAAAACU (((..((...((((((........)))))).))..)))...(((..((((.(((..(.((((((((...((((.....)))))))))))).)....))).)))).)))............ (-33.00 = -33.08 + 0.08)

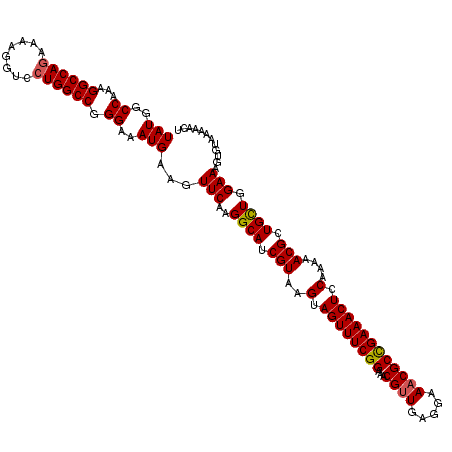
| Location | 11,383,896 – 11,384,016 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.20 |
| Mean single sequence MFE | -34.54 |
| Consensus MFE | -29.98 |
| Energy contribution | -30.98 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.87 |
| SVM decision value | 2.38 |
| SVM RNA-class probability | 0.993169 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
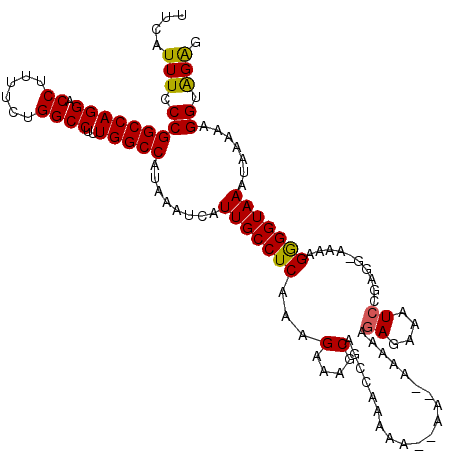
>3R_DroMel_CAF1 11383896 120 - 27905053 UUUUAUUUUUUUUUUGGCUGCUUUCUUUGAGGCAAUGAUUUAUGGCCAAAGGCCAAAAAAGGUCCUGGCCGGGAAAUGAAGUUCAAGGCAUCGUAAGUAGUUUCGGAAACGUUGAGGAAA ........(((((((((((((((....(((.((..((((((((..((...(((((..........))))).))..)))))..)))..)).))).)))))))).((....))..))))))) ( -31.30) >DroSec_CAF1 131623 116 - 1 CCUU--UU--UUUUUGGCUGCUUUCUUUGAGGCAAUGAUUUAUGGCCAAAGGCCAGAAAAGGUCCUGGCCGGGAAAUGAAGUUCAAGGCAUCGUAAGUAGUUUCGGAAACGAUGAGGAAA ((((--..--.....((((((((....(((.((..((((((((..((...((((((........)))))).))..)))))..)))..)).))).))))))))(((....))).))))... ( -39.30) >DroSim_CAF1 122724 118 - 1 UCUU--UUUUUUUUUGGCUGCUUUCUUUGAGGCAAUGAUUUAUGGCCAAAGGCCAGAAAAGGUCCUGGCCGGGAAAUGAAGUUCAAGGCAUCGUAAGUAGUUUCGGAAACGUUGAGGAAA ((((--(........((((((((....(((.((..((((((((..((...((((((........)))))).))..)))))..)))..)).))).)))))))).((....))..))))).. ( -36.20) >DroEre_CAF1 119095 109 - 1 UUU-------U---UUGCAG-CUUCUUUGAGGCAAUGAUUUAUGGCCAGAGGCCAGAAAAGGUCCUGGCCGGGAAAUGAAGUUCAAGGCAUCGUAAGUAGUUUCGGAAACGUUGAGGAAA .((-------(---(..(((-(...((((((((.....(((.(((((((.((((......))))))))))).)))((((.((.....)).)))).....))))))))...))))..)))) ( -33.80) >DroYak_CAF1 121842 113 - 1 UUU-------UUUUUUGCUGCCUUCUUUGAGGCAAUGAUUUAUGGCCAAAGGCCAGAAAAGGUCCUGGCCGGGAAAUGAAGUUCAAGGCAUCGUAAGUAGUUUCGGAAACGUUGAGGAAA .((-------((((((((((((((....)))))..((((((((..((...((((((........)))))).))..)))))..))).)))).............((....))..))))))) ( -32.10) >consensus UUUU__UU__UUUUUGGCUGCUUUCUUUGAGGCAAUGAUUUAUGGCCAAAGGCCAGAAAAGGUCCUGGCCGGGAAAUGAAGUUCAAGGCAUCGUAAGUAGUUUCGGAAACGUUGAGGAAA ................(((((((....(((.((..((((((((..((...((((((........)))))).))..)))))..)))..)).))).)))))))..((....))......... (-29.98 = -30.98 + 1.00)


| Location | 11,383,936 – 11,384,055 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.26 |
| Mean single sequence MFE | -28.07 |
| Consensus MFE | -21.31 |
| Energy contribution | -20.83 |
| Covariance contribution | -0.48 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.78 |
| SVM RNA-class probability | 0.847621 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
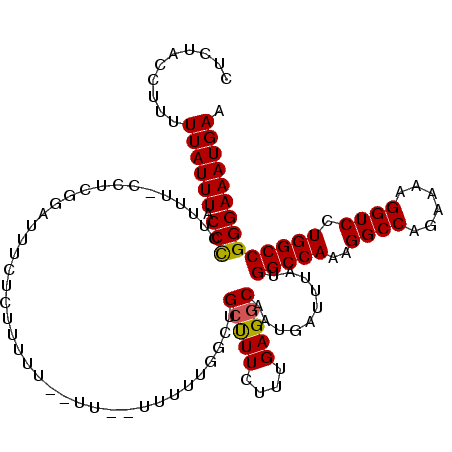
>3R_DroMel_CAF1 11383936 119 + 27905053 UUCAUUUCCCGGCCAGGACCUUUUUUGGCCUUUGGCCAUAAAUCAUUGCCUCAAAGAAAGCAGCCAAAAAAAAAAUAAAAAGAGAAAUACGAGA-AAAAGAGGUAAAUAAAAAGGUAGAG ....(((.(((((((((.((......))))..)))))........(((((((..........................................-....))))))).......)).))). ( -21.75) >DroSec_CAF1 131663 115 + 1 UUCAUUUCCCGGCCAGGACCUUUUCUGGCCUUUGGCCAUAAAUCAUUGCCUCAAAGAAAGCAGCCAAAAA--AA--AAGGAGAGAAAUCCGAGA-AAAAGGGGUAAAUAAAAAGGUAGAG ......((((((((((((....))))))))((((((......((.(((...))).)).....))))))..--..--..(((......)))....-....))))................. ( -27.50) >DroSim_CAF1 122764 117 + 1 UUCAUUUCCCGGCCAGGACCUUUUCUGGCCUUUGGCCAUAAAUCAUUGCCUCAAAGAAAGCAGCCAAAAAAAAA--AAGAAGAGAAAUCCGAGG-AAAAGGGGUAAAUAAAAAGGUAGAG ......((((((((((((....))))))))((((((......((.(((...))).)).....))))))......--.....((....)).....-....))))................. ( -25.10) >DroEre_CAF1 119135 109 + 1 UUCAUUUCCCGGCCAGGACCUUUUCUGGCCUCUGGCCAUAAAUCAUUGCCUCAAAGAAG-CUGCAA---A-------AAAAGAGAAAUCCGAGGCAAAAGGGGUAAAUAAGGAGGUGGGG (((((((((.(((((((.((......)).).))))))........(((((((...(...-...)..---.-------....((....)).))))))).............))))))))). ( -35.10) >DroYak_CAF1 121882 112 + 1 UUCAUUUCCCGGCCAGGACCUUUUCUGGCCUUUGGCCAUAAAUCAUUGCCUCAAAGAAGGCAGCAAAAAA-------AAAAGAGAAAUCCGAGG-AAACGGGGUAAAUAAAAAGGUAGAG ......(((((.((.(((..((((((((((...))))........((((((......)))))).......-------...)))))).)))..))-...)))))................. ( -30.90) >consensus UUCAUUUCCCGGCCAGGACCUUUUCUGGCCUUUGGCCAUAAAUCAUUGCCUCAAAGAAAGCAGCCAAAAA__AA__AAAAAGAGAAAUCCGAGG_AAAAGGGGUAAAUAAAAAGGUAGAG ....(((.(((((((((.((......))))..)))))........(((((((...............................................))))))).......)).))). (-21.31 = -20.83 + -0.48)


| Location | 11,383,936 – 11,384,055 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.26 |
| Mean single sequence MFE | -30.98 |
| Consensus MFE | -24.14 |
| Energy contribution | -23.94 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.79 |
| Structure conservation index | 0.78 |
| SVM decision value | 2.15 |
| SVM RNA-class probability | 0.989086 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11383936 119 - 27905053 CUCUACCUUUUUAUUUACCUCUUUU-UCUCGUAUUUCUCUUUUUAUUUUUUUUUUGGCUGCUUUCUUUGAGGCAAUGAUUUAUGGCCAAAGGCCAAAAAAGGUCCUGGCCGGGAAAUGAA .........................-.....((((((((...................((((((....)))))).........(((((..((((......)))).))))))))))))).. ( -25.70) >DroSec_CAF1 131663 115 - 1 CUCUACCUUUUUAUUUACCCCUUUU-UCUCGGAUUUCUCUCCUU--UU--UUUUUGGCUGCUUUCUUUGAGGCAAUGAUUUAUGGCCAAAGGCCAGAAAAGGUCCUGGCCGGGAAAUGAA .........(((((((.(((.....-....(((......)))..--..--..((((((((((((....)))))..........)))))))((((((........)))))))))))))))) ( -30.80) >DroSim_CAF1 122764 117 - 1 CUCUACCUUUUUAUUUACCCCUUUU-CCUCGGAUUUCUCUUCUU--UUUUUUUUUGGCUGCUUUCUUUGAGGCAAUGAUUUAUGGCCAAAGGCCAGAAAAGGUCCUGGCCGGGAAAUGAA .........(((((((.(((.....-..................--......((((((((((((....)))))..........)))))))((((((........)))))))))))))))) ( -29.10) >DroEre_CAF1 119135 109 - 1 CCCCACCUCCUUAUUUACCCCUUUUGCCUCGGAUUUCUCUUUU-------U---UUGCAG-CUUCUUUGAGGCAAUGAUUUAUGGCCAGAGGCCAGAAAAGGUCCUGGCCGGGAAAUGAA ..........((((((.(((...((((((((((...(((....-------.---..).))-....))))))))))........((((((.((((......))))))))))))))))))). ( -37.20) >DroYak_CAF1 121882 112 - 1 CUCUACCUUUUUAUUUACCCCGUUU-CCUCGGAUUUCUCUUUU-------UUUUUUGCUGCCUUCUUUGAGGCAAUGAUUUAUGGCCAAAGGCCAGAAAAGGUCCUGGCCGGGAAAUGAA ....................(((((-((.(((......(((((-------((..((..((((((....))))))..))....(((((...))))))))))))......)))))))))).. ( -32.10) >consensus CUCUACCUUUUUAUUUACCCCUUUU_CCUCGGAUUUCUCUUUUU__UU__UUUUUGGCUGCUUUCUUUGAGGCAAUGAUUUAUGGCCAAAGGCCAGAAAAGGUCCUGGCCGGGAAAUGAA ..........((((((.(((.......................................(((((....)))))..........(((((..((((......)))).)))))))))))))). (-24.14 = -23.94 + -0.20)

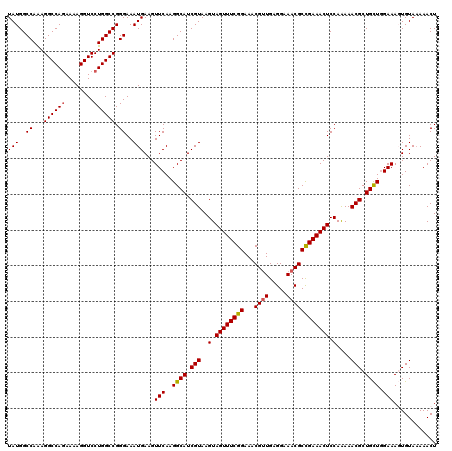
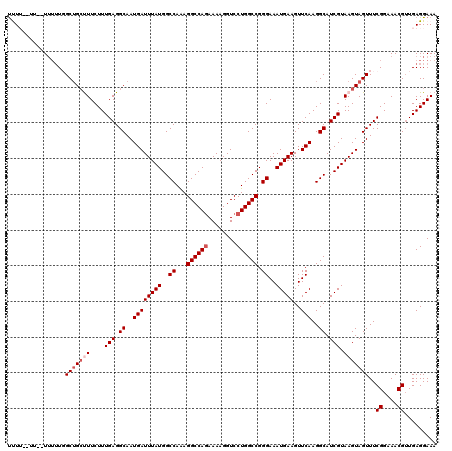
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:23:24 2006