
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 11,383,188 – 11,383,336 |
| Length | 148 |
| Max. P | 0.994195 |

| Location | 11,383,188 – 11,383,296 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.15 |
| Mean single sequence MFE | -37.20 |
| Consensus MFE | -31.89 |
| Energy contribution | -31.73 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.86 |
| SVM decision value | 2.46 |
| SVM RNA-class probability | 0.994195 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11383188 108 + 27905053 AUUGGCCUGACUGGCGAUU------------CGCAUUCGCUGGAUUGCGUGUCUUCGCCAAUUUGUUGGGAGAUUAAACAAAUUUCCUGCCAACGGCCACGCAAUUUAUACCCAUAUACG ..(((((....((((((..------------((((.((....)).)))).....))))))..(((..((((((((.....))))))))..))).)))))..................... ( -34.70) >DroSec_CAF1 130910 108 + 1 AUUGGCCUGACUGGCGAUU------------CGCAUUCGCUGGAUUGCGUGUCUUCGCCAAUUUGUUGGGAGAUUAAACAAAUUUCCUGCCAACGGCCACGCAAUUUGUGCCUAUAUACG ..(((((....((((((..------------((((.((....)).)))).....))))))..(((..((((((((.....))))))))..))).))))).(((.....)))......... ( -36.00) >DroSim_CAF1 122034 108 + 1 AUUGGCCUGACUGGCGAUU------------CGCAUUCGCUGGAUUGCGUGUCUUCGCCAAUUUGUUGGGAGAUUAAACAAAUUUCCUGCCAACGGCCACGCAAUUUGUGCCCAUAUACG ..(((((....((((((..------------((((.((....)).)))).....))))))..(((..((((((((.....))))))))..))).))))).(((.....)))......... ( -36.00) >DroEre_CAF1 118399 120 + 1 AUUGGCCUGACUGGCGAUUCGGAUUCGGAUUCGCAUUCGGUGGAUUGCGUGUCUUCGCCAAUUUGUUGGGAGAUUAAACAAAUUUCCUGCCAGCGGCCACGCAAUUUGUGCCCAUAUACG ..(((((.(.(((((.....((....((((.((((.((....)).)))).))))...))........((((((((.....))))))))))))))))))).(((.....)))......... ( -39.70) >DroYak_CAF1 121059 114 + 1 AUUGGCCUGACUGGCGAUUCG------GAUUCGCAUUCGGCGGAUUGCGUGUCUUCGCCAAUUUGUUGGGAGAUUAUACAAAUUUCCUGCCAACGGCCACGCAAUUUGUGCCCAUAUACG ..(((((....((((((...(------(((.((((.((....)).)))).))))))))))..(((..((((((((.....))))))))..))).))))).(((.....)))......... ( -39.60) >consensus AUUGGCCUGACUGGCGAUU____________CGCAUUCGCUGGAUUGCGUGUCUUCGCCAAUUUGUUGGGAGAUUAAACAAAUUUCCUGCCAACGGCCACGCAAUUUGUGCCCAUAUACG ..(((((....((((((..............((((.((....)).)))).....))))))..(((..((((((((.....))))))))..))).)))))..................... (-31.89 = -31.73 + -0.16)

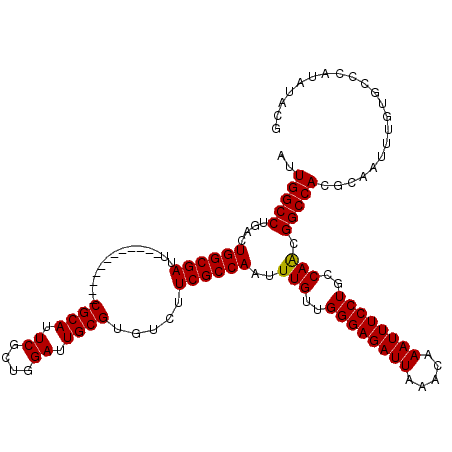
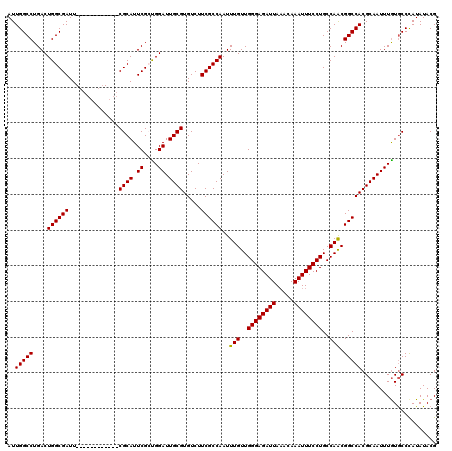
| Location | 11,383,188 – 11,383,296 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.15 |
| Mean single sequence MFE | -33.52 |
| Consensus MFE | -29.43 |
| Energy contribution | -29.47 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.47 |
| SVM RNA-class probability | 0.956670 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11383188 108 - 27905053 CGUAUAUGGGUAUAAAUUGCGUGGCCGUUGGCAGGAAAUUUGUUUAAUCUCCCAACAAAUUGGCGAAGACACGCAAUCCAGCGAAUGCG------------AAUCGCCAGUCAGGCCAAU ((((.............))))((((((((((.(((............))).)))))..((((((((.....((((.((....)).))))------------..))))))))..))))).. ( -31.82) >DroSec_CAF1 130910 108 - 1 CGUAUAUAGGCACAAAUUGCGUGGCCGUUGGCAGGAAAUUUGUUUAAUCUCCCAACAAAUUGGCGAAGACACGCAAUCCAGCGAAUGCG------------AAUCGCCAGUCAGGCCAAU .........(((.....))).((((((((((.(((............))).)))))..((((((((.....((((.((....)).))))------------..))))))))..))))).. ( -32.80) >DroSim_CAF1 122034 108 - 1 CGUAUAUGGGCACAAAUUGCGUGGCCGUUGGCAGGAAAUUUGUUUAAUCUCCCAACAAAUUGGCGAAGACACGCAAUCCAGCGAAUGCG------------AAUCGCCAGUCAGGCCAAU ((((..((....))...))))((((((((((.(((............))).)))))..((((((((.....((((.((....)).))))------------..))))))))..))))).. ( -33.40) >DroEre_CAF1 118399 120 - 1 CGUAUAUGGGCACAAAUUGCGUGGCCGCUGGCAGGAAAUUUGUUUAAUCUCCCAACAAAUUGGCGAAGACACGCAAUCCACCGAAUGCGAAUCCGAAUCCGAAUCGCCAGUCAGGCCAAU ((((..((....))...))))(((((((((((.(((....(((((....((((((....)))).)))))))((((.((....)).))))..)))((.......))))))))..))))).. ( -34.90) >DroYak_CAF1 121059 114 - 1 CGUAUAUGGGCACAAAUUGCGUGGCCGUUGGCAGGAAAUUUGUAUAAUCUCCCAACAAAUUGGCGAAGACACGCAAUCCGCCGAAUGCGAAUC------CGAAUCGCCAGUCAGGCCAAU ((((..((....))...))))((((((((((.(((..((....))..))).)))))..((((((((.((..((((.((....)).))))..))------....))))))))..))))).. ( -34.70) >consensus CGUAUAUGGGCACAAAUUGCGUGGCCGUUGGCAGGAAAUUUGUUUAAUCUCCCAACAAAUUGGCGAAGACACGCAAUCCAGCGAAUGCG____________AAUCGCCAGUCAGGCCAAU .........(((.....))).((((((((((.(((............))).)))))..((((((((.....((((.((....)).))))..............))))))))..))))).. (-29.43 = -29.47 + 0.04)



| Location | 11,383,216 – 11,383,336 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.67 |
| Mean single sequence MFE | -37.76 |
| Consensus MFE | -35.26 |
| Energy contribution | -35.18 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.24 |
| SVM RNA-class probability | 0.935125 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11383216 120 + 27905053 UGGAUUGCGUGUCUUCGCCAAUUUGUUGGGAGAUUAAACAAAUUUCCUGCCAACGGCCACGCAAUUUAUACCCAUAUACGCGACCUACAGGUUGGGAUCCUCGACUUUUGUCUGGAUGUC (((((((((((.....(((...(((..((((((((.....))))))))..))).))))))))))))))..........(.(((((....))))).)((((..(((....))).))))... ( -39.90) >DroSec_CAF1 130938 120 + 1 UGGAUUGCGUGUCUUCGCCAAUUUGUUGGGAGAUUAAACAAAUUUCCUGCCAACGGCCACGCAAUUUGUGCCUAUAUACGCGACCUACAGGUGGGGAUCCUCGACUUUUGUCUGGAUGUC .((((((((((.....(((...(((..((((((((.....))))))))..))).)))))))))))))...........(.(.(((....))).).)((((..(((....))).))))... ( -37.50) >DroSim_CAF1 122062 120 + 1 UGGAUUGCGUGUCUUCGCCAAUUUGUUGGGAGAUUAAACAAAUUUCCUGCCAACGGCCACGCAAUUUGUGCCCAUAUACGCGACCUACAGGUGGGGAUCCUCGACUUUUGUCUGGAUGUC .((((((((((.....(((...(((..((((((((.....))))))))..))).)))))))))))))...........(.(.(((....))).).)((((..(((....))).))))... ( -37.50) >DroEre_CAF1 118439 120 + 1 UGGAUUGCGUGUCUUCGCCAAUUUGUUGGGAGAUUAAACAAAUUUCCUGCCAGCGGCCACGCAAUUUGUGCCCAUAUACGCGACCUACAGGUGGGGAUCCUCGACUUUUGUCUGGCUGUC .((((((((((.....(((...(((..((((((((.....))))))))..))).)))))))))))))..(((......(.(.(((....))).).)......(((....))).))).... ( -37.10) >DroYak_CAF1 121093 120 + 1 CGGAUUGCGUGUCUUCGCCAAUUUGUUGGGAGAUUAUACAAAUUUCCUGCCAACGGCCACGCAAUUUGUGCCCAUAUACGCGACCUACAGGUGGGGAUCCUCUACUCUUGUCUGGAUGUC (((((((((((.....(((...(((..((((((((.....))))))))..))).)))))))))))))).((........))(((...((((..(((.........)))..))))...))) ( -36.80) >consensus UGGAUUGCGUGUCUUCGCCAAUUUGUUGGGAGAUUAAACAAAUUUCCUGCCAACGGCCACGCAAUUUGUGCCCAUAUACGCGACCUACAGGUGGGGAUCCUCGACUUUUGUCUGGAUGUC (((((((((((.....(((...(((..((((((((.....))))))))..))).))))))))))))))..........(.(.(((....))).).)((((..(((....))).))))... (-35.26 = -35.18 + -0.08)

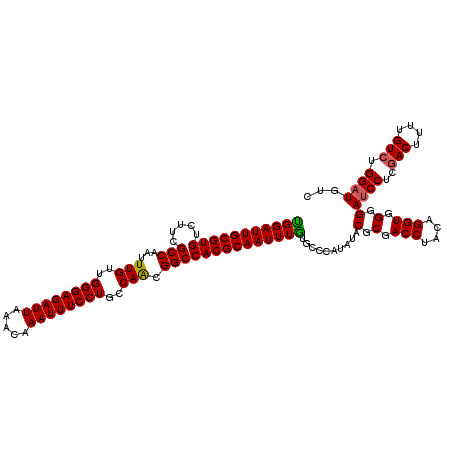

| Location | 11,383,216 – 11,383,336 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.67 |
| Mean single sequence MFE | -34.21 |
| Consensus MFE | -32.14 |
| Energy contribution | -32.62 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.11 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.00 |
| SVM RNA-class probability | 0.535365 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11383216 120 - 27905053 GACAUCCAGACAAAAGUCGAGGAUCCCAACCUGUAGGUCGCGUAUAUGGGUAUAAAUUGCGUGGCCGUUGGCAGGAAAUUUGUUUAAUCUCCCAACAAAUUGGCGAAGACACGCAAUCCA ...((((.(((....)))..)))).....(((((.(((((((((.............)))))))))....))))).((((((((.........)))))))).(((......)))...... ( -33.82) >DroSec_CAF1 130938 120 - 1 GACAUCCAGACAAAAGUCGAGGAUCCCCACCUGUAGGUCGCGUAUAUAGGCACAAAUUGCGUGGCCGUUGGCAGGAAAUUUGUUUAAUCUCCCAACAAAUUGGCGAAGACACGCAAUCCA ...((((.(((....)))..)))).....(((((.(((((((((.............)))))))))....))))).((((((((.........)))))))).(((......)))...... ( -33.82) >DroSim_CAF1 122062 120 - 1 GACAUCCAGACAAAAGUCGAGGAUCCCCACCUGUAGGUCGCGUAUAUGGGCACAAAUUGCGUGGCCGUUGGCAGGAAAUUUGUUUAAUCUCCCAACAAAUUGGCGAAGACACGCAAUCCA ...((((.(((....)))..)))).....(((((.(((((((((..((....))...)))))))))....))))).((((((((.........)))))))).(((......)))...... ( -35.40) >DroEre_CAF1 118439 120 - 1 GACAGCCAGACAAAAGUCGAGGAUCCCCACCUGUAGGUCGCGUAUAUGGGCACAAAUUGCGUGGCCGCUGGCAGGAAAUUUGUUUAAUCUCCCAACAAAUUGGCGAAGACACGCAAUCCA ....(((((.(........(((.......)))...(((((((((..((....))...))))))))))))))).(((((((((((.........)))))))).(((......)))..))). ( -35.90) >DroYak_CAF1 121093 120 - 1 GACAUCCAGACAAGAGUAGAGGAUCCCCACCUGUAGGUCGCGUAUAUGGGCACAAAUUGCGUGGCCGUUGGCAGGAAAUUUGUAUAAUCUCCCAACAAAUUGGCGAAGACACGCAAUCCG ....................((((.....(((((.(((((((((..((....))...)))))))))....))))).(((((((...........))))))).(((......))).)))). ( -32.10) >consensus GACAUCCAGACAAAAGUCGAGGAUCCCCACCUGUAGGUCGCGUAUAUGGGCACAAAUUGCGUGGCCGUUGGCAGGAAAUUUGUUUAAUCUCCCAACAAAUUGGCGAAGACACGCAAUCCA ........(((....)))..((((.....(((((.(((((((((..((....))...)))))))))....))))).((((((((.........)))))))).(((......))).)))). (-32.14 = -32.62 + 0.48)

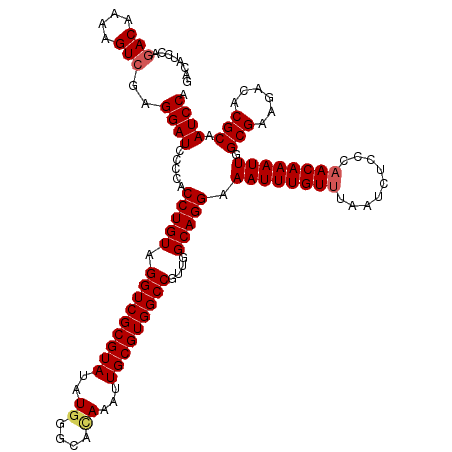

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:23:19 2006