
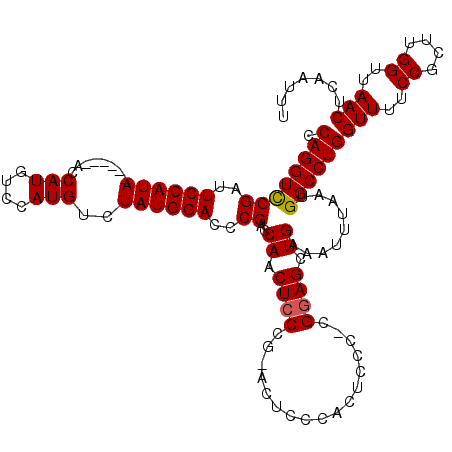
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 11,381,520 – 11,381,663 |
| Length | 143 |
| Max. P | 0.759017 |

| Location | 11,381,520 – 11,381,633 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.82 |
| Mean single sequence MFE | -26.53 |
| Consensus MFE | -20.27 |
| Energy contribution | -20.23 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.76 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.507970 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11381520 113 - 27905053 CCAGGUCCGAUUCCAUA------ACCAUGUCCAUGUCUAUGGACCCGACCAACUCCCGAACUCCCACUACC-CGGAGCUGAAAUUUAAUGGCCUGGUUUUCCGCUUGGUUAACUCAAUUU .((((((((..((((((------..(((....)))..))))))..))..((.((((.(............)-.)))).)).........))))))(((..((....))..)))....... ( -27.30) >DroSec_CAF1 129283 113 - 1 CCAGGUUCGAUUCCAUA------ACCAUGUCCAUGUCUAUGGAGCCGACCAACUCCCGCACUCCCACUCCC-CGGAGCUGAAAUUUAAUGGCCUGGUUUUCCGCUUGGUUAACUCAAUUU .((((.(((..((((((------..(((....)))..))))))..)))(((.......((((((.......-.)))).))........)))))))(((..((....))..)))....... ( -27.86) >DroSim_CAF1 120337 113 - 1 GCAGGUUCGAUUCCAUA------ACCAUGUCCAUGUCUAUGGAGCCGACCAACUCCCGAACUCCCACUCCC-CGUAGCUGAAAUUUAAUGGCCUGGUUUUCCGCUUGGUUAACUCAAUUU ...(((.((..((((((------..(((....)))..))))))..)))))....................(-((.(((.((((.(((......))).)))).))))))............ ( -23.60) >DroEre_CAF1 116769 112 - 1 CCAGGUCCGUUUCCAUAACCAUAACCAUGUCCAUGUCUAUGGACCCGACCAACUCCC--------ACUCCCGCGGAGCUGAAAUUUAAUGGCCUGGUUUUCCGCUUGGUUAACUCAAUUU .((((((((..((((((..(((..........)))..))))))..))..((.(((((--------......).)))).)).........))))))(((..((....))..)))....... ( -27.60) >DroYak_CAF1 119375 118 - 1 CCAGGUCCGAUUCCAUAACCAUAACCAUGUCCAUGUCUAUGGACCCGACCAACUCCC--ACUCCCACUCCCGCGGAGCUGAAAUUUAAUGGCCUGGUUUUCCGCUUGGUUAACUCAAUUU ((((((..((..(((...((((......((((((....))))))............(--((((((......).)))).)).......))))..)))..))..))))))............ ( -26.30) >consensus CCAGGUCCGAUUCCAUA______ACCAUGUCCAUGUCUAUGGACCCGACCAACUCCCG_ACUCCCACUCCC_CGGAGCUGAAAUUUAAUGGCCUGGUUUUCCGCUUGGUUAACUCAAUUU .((((((((..((((((........(((....)))..))))))..))..((.((((.................)))).)).........))))))(((..((....))..)))....... (-20.27 = -20.23 + -0.04)

| Location | 11,381,560 – 11,381,663 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.81 |
| Mean single sequence MFE | -42.60 |
| Consensus MFE | -27.57 |
| Energy contribution | -27.93 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.759017 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11381560 103 + 27905053 CAGCUCCG-GGUAGUGGGAGUUCGGGAGUUGGUCGGGUCCAUAGACAUGGACAUGGU------UAUGGAAUCGGACCUGGAGCAGAGGCAACUCCACUUCAGCCGCUCC----------G .((((((.-.......))))))..(((((.((((.(((((((((.(((....))).)------))))).))).))))(((((..(((....)))..)))))...)))))----------. ( -42.40) >DroSec_CAF1 129323 103 + 1 CAGCUCCG-GGGAGUGGGAGUGCGGGAGUUGGUCGGCUCCAUAGACAUGGACAUGGU------UAUGGAAUCGAACCUGGAGCAGAGGCAACUCCACUUCAGCCGCUCC----------G ..(((((.-.......)))))...(((((.(((((..(((((((.(((....))).)------))))))..)))..((((((..(((....)))..)))))))))))))----------. ( -43.10) >DroSim_CAF1 120377 103 + 1 CAGCUACG-GGGAGUGGGAGUUCGGGAGUUGGUCGGCUCCAUAGACAUGGACAUGGU------UAUGGAAUCGAACCUGCAGCAGAGGCAACUCCACUUCAGCCGCUCC----------G ...(....-)(((((((..(....(((((((.(((..(((((((.(((....))).)------))))))..))).(((.......))))))))))....)..)))))))----------. ( -41.00) >DroEre_CAF1 116809 112 + 1 CAGCUCCGCGGGAGU--------GGGAGUUGGUCGGGUCCAUAGACAUGGACAUGGUUAUGGUUAUGGAAACGGACCUGGAGUAGAGGGAACUCCACUUCAGCCGCUCCGCUCCGCUCCG .......((((.(((--------((.(((.(((((..(((((((.(((((......))))).)))))))..)))..(((((((.(((....))).))))))))))))))))))))).... ( -47.30) >DroYak_CAF1 119415 100 + 1 CAGCUCCGCGGGAGUGGGAGU--GGGAGUUGGUCGGGUCCAUAGACAUGGACAUGGUUAUGGUUAUGGAAUCGGACCUGGAGUAGAGGGAACUCCACUUCA------------------G ..(((((((....)).)))))--.......((((.(((((((((.(((((......))))).)))))).))).))))((((((.(((....))).))))))------------------. ( -39.20) >consensus CAGCUCCG_GGGAGUGGGAGU_CGGGAGUUGGUCGGGUCCAUAGACAUGGACAUGGU______UAUGGAAUCGGACCUGGAGCAGAGGCAACUCCACUUCAGCCGCUCC__________G (((((((.................))))))).(((..((((((..(((....)))........))))))..)))..((((((..(((....)))..)))))).................. (-27.57 = -27.93 + 0.36)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:23:15 2006