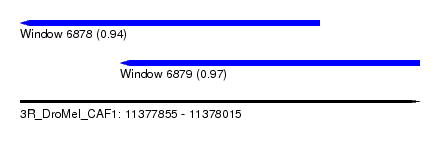
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 11,377,855 – 11,378,015 |
| Length | 160 |
| Max. P | 0.971006 |

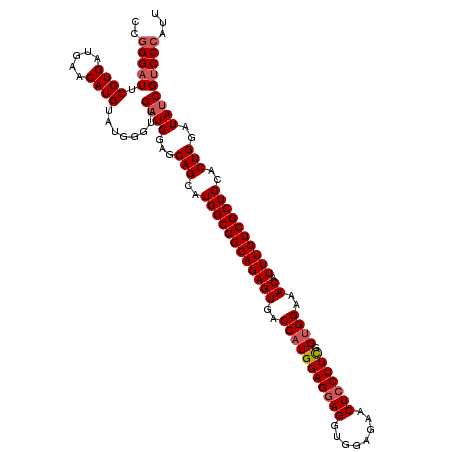
| Location | 11,377,855 – 11,377,975 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.50 |
| Mean single sequence MFE | -44.56 |
| Consensus MFE | -40.48 |
| Energy contribution | -40.88 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.31 |
| SVM RNA-class probability | 0.940996 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11377855 120 - 27905053 AUGGCGGCAGAGUGACCAUGGAGGAGGUGGAGAACUCCUUCGGCGGAAACAUUUGUCGCUGCACUGGAUAUCGUCCCAUUCUGGAUGCCAUGAAGUCCUUUGCGGUGGACAGCACCGUUG ..(((((((((((..((.((((((((........))))))))..))..)).)))))))))(((..((((.((((..((((...))))..)))).))))..)))((((.....)))).... ( -47.20) >DroSec_CAF1 125497 120 - 1 AUGGCGGCAGAGUGACCAUGGAGGAGGUGGAGAACUCCUUCGGUGGAAACAUUUGUCGCUGCACUGGAUAUCGUCCCAUUCUGGAUGCCAUGAAGUCCUUUGCGGUGGACAGCACCAUUG ..(((((((((((..(((((((((((........))))))).))))..)).)))))))))(((..((((.((((..((((...))))..)))).))))..)))((((.....)))).... ( -48.30) >DroSim_CAF1 116501 120 - 1 AUGGCGGCAGAGUGACCAUGGAGGAGGUGGAGAACUCCUUUGGUGGAAACAUUUGUCGCUGCACUGGAUAUCGUCCAAUUCUGGAUGCCAUGAAGUCCUUUGCUGUGGACAGCACCAUUG ..(((((((((((..((((.((((((........)))).)).))))..)).)))))))))((..(((....((((((....)))))))))....((((........)))).))....... ( -43.20) >DroEre_CAF1 113050 120 - 1 AUGGCGGCAGAGUGACCAUGGAGCAGGUGGAGAACUCCUUCGGUGGAAACAUUUGUCGCUGCACUGGAUACCGUCCCAUUCUGGAUGCCAUGAAGUCCUUUGCGGUGGACAGCACUGUUG ...((.((((((.((((((((.((((..(((....)))...((((....)))).....)))).........(((((......))))))))))..))))))))).)).((((....)))). ( -39.60) >DroYak_CAF1 115590 120 - 1 AUGGCGGCAGAGUGACCAUGGAGGAGGUGGAGAACUCCUUUGGUGGAAACAUUUGUCGCUGCACUGGAUAUCGGCCCAUUCUGGAUGCCAUGAAGUCCUUUGCGGUGGACAGCACUAUUG ..(((((((((((..((((.((((((........)))).)).))))..)).)))))))))(((..((((.(((((((.....))..)))..)).))))..)))((((.....)))).... ( -44.50) >consensus AUGGCGGCAGAGUGACCAUGGAGGAGGUGGAGAACUCCUUCGGUGGAAACAUUUGUCGCUGCACUGGAUAUCGUCCCAUUCUGGAUGCCAUGAAGUCCUUUGCGGUGGACAGCACCAUUG ..(((((((((((..(((((((((((........))))))).))))..)).)))))))))(((..((((..(((((......))))).......))))..)))((((.....)))).... (-40.48 = -40.88 + 0.40)

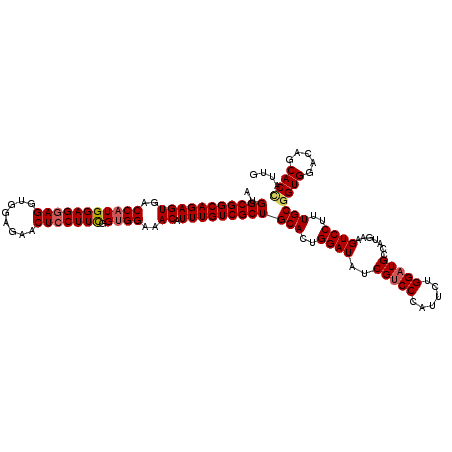
| Location | 11,377,895 – 11,378,015 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.50 |
| Mean single sequence MFE | -43.76 |
| Consensus MFE | -38.52 |
| Energy contribution | -39.28 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.67 |
| SVM RNA-class probability | 0.971006 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11377895 120 - 27905053 CCGGGAUUCGUGAUGAACAUGUAUGGGUUGAUGGAACAGCAUGGCGGCAGAGUGACCAUGGAGGAGGUGGAGAACUCCUUCGGCGGAAACAUUUGUCGCUGCACUGGAUAUCGUCCCAUU ........((((.....)))).(((((.(((((...(((..((((((((((((..((.((((((((........))))))))..))..)).))))))))))..)))..))))).))))). ( -45.10) >DroSec_CAF1 125537 120 - 1 CCGGGAUUCGUGAUGAACAUGUAUGGGUUGAUGGAGCAGCAUGGCGGCAGAGUGACCAUGGAGGAGGUGGAGAACUCCUUCGGUGGAAACAUUUGUCGCUGCACUGGAUAUCGUCCCAUU ........((((.....)))).(((((.(((((...(((..((((((((((((..(((((((((((........))))))).))))..)).))))))))))..)))..))))).))))). ( -45.10) >DroSim_CAF1 116541 120 - 1 CCGGGAUUCGUGAUGAACAUGUAUGGGUUGAUGGAGCAGCAUGGCGGCAGAGUGACCAUGGAGGAGGUGGAGAACUCCUUUGGUGGAAACAUUUGUCGCUGCACUGGAUAUCGUCCAAUU ...((((....((((..(((((....((((......))))..(((((((((((..((((.((((((........)))).)).))))..)).)))))))))))).))..)))))))).... ( -38.80) >DroEre_CAF1 113090 120 - 1 CCGGGAUUCGUGAUGAACAUGUAUGGCUUGAUGGAGCAGCAUGGCGGCAGAGUGACCAUGGAGCAGGUGGAGAACUCCUUCGGUGGAAACAUUUGUCGCUGCACUGGAUACCGUCCCAUU ..(((((..(((.....(((((...((((....)))).)))))((((((((((..((((.(((..(((.....)))..))).))))..)).))))))))..))).((...)))))))... ( -41.60) >DroYak_CAF1 115630 120 - 1 CCGGGAUUCGUGAUGAACAUGUACGGGCUGAUGGAGCAGCAUGGCGGCAGAGUGACCAUGGAGGAGGUGGAGAACUCCUUUGGUGGAAACAUUUGUCGCUGCACUGGAUAUCGGCCCAUU ........((((.....))))...(((((((((...(((..((((((((((((..((((.((((((........)))).)).))))..)).))))))))))..)))..)))))))))... ( -48.20) >consensus CCGGGAUUCGUGAUGAACAUGUAUGGGUUGAUGGAGCAGCAUGGCGGCAGAGUGACCAUGGAGGAGGUGGAGAACUCCUUCGGUGGAAACAUUUGUCGCUGCACUGGAUAUCGUCCCAUU ..(((((.((((.....))))........((((...(((..((((((((((((..(((((((((((........))))))).))))..)).))))))))))..)))..)))))))))... (-38.52 = -39.28 + 0.76)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:23:12 2006