
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 11,376,298 – 11,376,618 |
| Length | 320 |
| Max. P | 0.999410 |

| Location | 11,376,298 – 11,376,418 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.50 |
| Mean single sequence MFE | -43.56 |
| Consensus MFE | -36.44 |
| Energy contribution | -36.68 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.754870 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11376298 120 - 27905053 GGUUACGAAGCGGAGGAGAUUUUCUGCUCUGGAAUGGUGCGCCACUCGGAACAACCAGUGGGUGUAAUCGUGGCUCUUACUGCUGACCAAGCCCAAAGGGCAACAAAGCUUGUCAAGAUC .(((((((..(((((.((.....)).)))))......(((((((((.((.....))))).)))))).)))))))((((((.(((......((((...)))).....)))..)).)))).. ( -39.40) >DroSec_CAF1 123937 120 - 1 GGUUACGAAGCGGAGGAGAUCUUCUGCUCUGGAACGGUGCGCCACUCCGAACAACCAGUGGGUGUAAUCGUGGCUCUUACUGCUGACCAAGCUCAAAGGGCCACAAAACUGGUCAAGAUC ((((((((.(((((((....)))))))))((((..((....))..))))....(((....)))))))))(((((((((...(((.....)))...)))))))))................ ( -42.30) >DroSim_CAF1 114941 120 - 1 GGUUACGAAGCGGAGGAGAUCUUCUGCUCUGGAAUGGUGCGCCACUCCGAACAGCCAGUGGGUGUAAUCGUGGCUCUUACUGCUGACCAAGCACAAAGGGCCACAAAACUGGUCAGGAUC ((((((((.(((((((....)))))))))((((.(((....))).))))....(((....)))))))))(((((((((..((((.....))))..)))))))))................ ( -46.20) >DroEre_CAF1 111492 120 - 1 GGCUACGAAGCCGAGGAGAUCUUUUGCUCCGGAUUGGUGCGCCACUCGGAACAGCCAGUGGGUGUAAUCGUGGCUCUCACUGCUGACCAAGCGCAAAGGGCCACUAAGUUGGUCAAGAUU ((((....))))..((((........)))).(((..((((((((((.((.....))))).))))))...((((((((...((((.....))))...))))))))....)..)))...... ( -43.50) >DroYak_CAF1 114034 120 - 1 GGCUACGAAGCCGAGGAGAUCUUCUGCUCGGGAUUGGUGCGCCACUCGGAGCAGCCAGUGGGUGUAAUCGUGGCUCUUACUGCUGACCAAGCCCAAAGGGCCACUAAGUUGGUCAAGAUU ((((((....(((((.((.....)).)))))((((.(..(.(((((.((.....))))))))..)))))(((((((((...(((.....)))...)))))))))...).)))))...... ( -46.40) >consensus GGUUACGAAGCGGAGGAGAUCUUCUGCUCUGGAAUGGUGCGCCACUCGGAACAGCCAGUGGGUGUAAUCGUGGCUCUUACUGCUGACCAAGCCCAAAGGGCCACAAAGCUGGUCAAGAUC ((((.((.((((((((....)))))))).))((...(..(.(((((.((.....))))))))..)..))(((((((((...(((.....)))...)))))))))..)))).......... (-36.44 = -36.68 + 0.24)



| Location | 11,376,418 – 11,376,538 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.42 |
| Mean single sequence MFE | -37.92 |
| Consensus MFE | -33.27 |
| Energy contribution | -32.75 |
| Covariance contribution | -0.52 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.31 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.728566 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11376418 120 - 27905053 AAGGUGGGAGCCACCAUUGAUCAAAUCGAUGCUACGGAAGCUCUCCAACAUCCAGGAGUGGUGGCCUUUUACACUGCGAAGGACAUUCCCGGAACGAAUACUUUCUGUGAGCCCAGUUUU (((.((((.((((((((((((...))))))(((.....))).((((........)))).))))))..(((((((((.(((.....))).)))...(((....))))))))))))).))). ( -33.10) >DroSec_CAF1 124057 120 - 1 AAAGUGGGAGCCACCAUCGAUCAAAUCGAUGCUUCGGAAGCUCUUCAACAUCCUGGAGUGGUGGCCUUCUACACUGCGAAGGACAUUCCCGGAACGAAUACUUUCUGUGAGCCCAGUUUU (((.((((...((((((((((...))))))).(((((((....)))....(((.((((((....(((((........))))).)))))).))).))))........)))..)))).))). ( -38.60) >DroSim_CAF1 115061 120 - 1 AAAGUGGGAGCCACCAUCGAUCAAAUCGAUGCUUCGGAAGCUCUCCAACAUCCAGGAGUGGUGGCCUUCUACACUGCGAAGGACAUUCCCGGAACGAAUACUUUCUGUGAGCCCAGUUUU (((.((((...((((((((((...))))))).((((((.....)))....(((.((((((....(((((........))))).)))))).)))..)))........)))..)))).))). ( -38.40) >DroEre_CAF1 111612 120 - 1 AAGGUGGGAGCCACCGUCGAUCAAAUCGAUGCUUCGGCAGCUCUCCAAUAUCCAGGAGUGGUGGCCUUUUACACUGCAAAGGACAUUCCCGGCACGAAUACUUUCUGUGAGCCCAGUUUU (((.((((.((((((((((((...))))))((....))....((((........)))).))))))..((((((.(((...((......)).))).(((....))))))))))))).))). ( -39.30) >DroYak_CAF1 114154 120 - 1 AAGGUGGGAGCCACCAUCGACCAAAUCGAUGCCUCGGAAGCUCUACAACAUCCAGGCGUGGUGGCCUUUUACACUGCGAAGGACAUUCCCGGAACGAAUACUUUCUGUGAGCCCAGCUUU (((.((((.(((((((((((.....)))))((((.(((.(........).)))))))..))))))..(((((((((.(((.....))).)))...(((....))))))))))))).))). ( -40.20) >consensus AAGGUGGGAGCCACCAUCGAUCAAAUCGAUGCUUCGGAAGCUCUCCAACAUCCAGGAGUGGUGGCCUUUUACACUGCGAAGGACAUUCCCGGAACGAAUACUUUCUGUGAGCCCAGUUUU (((.((((...(((((((((.....))))))....(((((..........(((.((((((....(((((........))))).)))))).))).......))))).)))..)))).))). (-33.27 = -32.75 + -0.52)

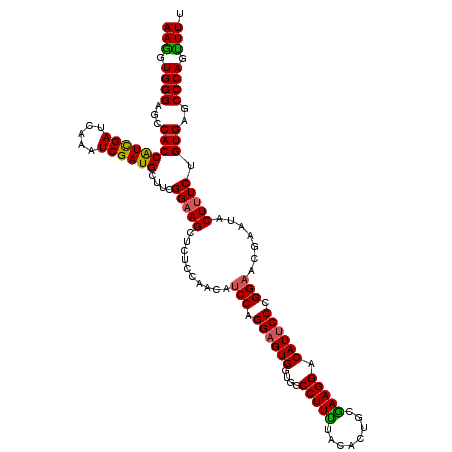
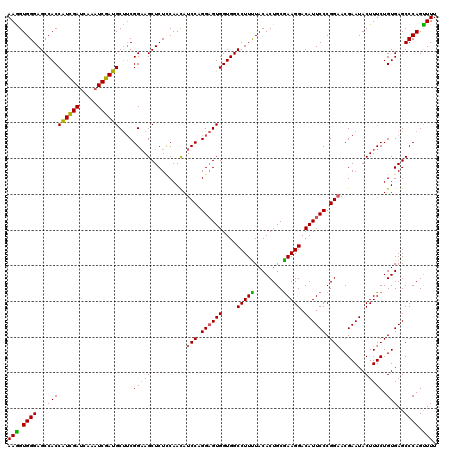
| Location | 11,376,458 – 11,376,578 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.58 |
| Mean single sequence MFE | -43.16 |
| Consensus MFE | -39.00 |
| Energy contribution | -39.24 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.21 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.699519 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11376458 120 + 27905053 UUCGCAGUGUAAAAGGCCACCACUCCUGGAUGUUGGAGAGCUUCCGUAGCAUCGAUUUGAUCAAUGGUGGCUCCCACCUUUGUGGCGCCCACAAAGGCGCAGUAAACGGAGUUGGAAGUG ..(((.........((((((((((((........)))).(((.....)))..............)))))))).(((.((((((.(((((......))))).....)))))).)))..))) ( -42.60) >DroSec_CAF1 124097 120 + 1 UUCGCAGUGUAGAAGGCCACCACUCCAGGAUGUUGAAGAGCUUCCGAAGCAUCGAUUUGAUCGAUGGUGGCUCCCACUUUCGUGGCGCCCACAAAGGCGCAGUAAACGGAGUUGGAAGUG ..(((.........(((((((......(((.(((....))).))).....((((((...))))))))))))).(((.((((((.(((((......))))).....)))))).)))..))) ( -41.80) >DroSim_CAF1 115101 120 + 1 UUCGCAGUGUAGAAGGCCACCACUCCUGGAUGUUGGAGAGCUUCCGAAGCAUCGAUUUGAUCGAUGGUGGCUCCCACUUUCGUGGCACCCACGAAGGCGCAGUAAACGGAGUUGGAAGUG ((((.(.(((.(..(((((((.((((........)))).(((.....)))((((((...)))))))))))))..).(((((((((...))))))))).))).)...)))).......... ( -39.40) >DroEre_CAF1 111652 120 + 1 UUUGCAGUGUAAAAGGCCACCACUCCUGGAUAUUGGAGAGCUGCCGAAGCAUCGAUUUGAUCGACGGUGGCUCCCACCUUUGUGGCGCCCACAAAGGCGCAGUAAACGGAGUUGGAAGUG ...((.........(((((((.((((........)))).(((.....))).(((((...))))).))))))).(((.((((((.(((((......))))).....)))))).)))..)). ( -43.70) >DroYak_CAF1 114194 120 + 1 UUCGCAGUGUAAAAGGCCACCACGCCUGGAUGUUGUAGAGCUUCCGAGGCAUCGAUUUGGUCGAUGGUGGCUCCCACCUUUGUGGCGCCCACAAAGGCGCAGUAGACGGAGUUGGAAGUG ..(((.........(((((((..(((((((.(((....))).))).))))((((((...))))))))))))).(((.((((((.(((((......))))).....)))))).)))..))) ( -48.30) >consensus UUCGCAGUGUAAAAGGCCACCACUCCUGGAUGUUGGAGAGCUUCCGAAGCAUCGAUUUGAUCGAUGGUGGCUCCCACCUUUGUGGCGCCCACAAAGGCGCAGUAAACGGAGUUGGAAGUG ..(((.........(((((((.((((........)))).(((.....)))((((((...))))))))))))).(((.((((((.(((((......))))).....)))))).)))..))) (-39.00 = -39.24 + 0.24)



| Location | 11,376,458 – 11,376,578 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.58 |
| Mean single sequence MFE | -42.34 |
| Consensus MFE | -38.54 |
| Energy contribution | -38.26 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.91 |
| SVM decision value | 3.58 |
| SVM RNA-class probability | 0.999410 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11376458 120 - 27905053 CACUUCCAACUCCGUUUACUGCGCCUUUGUGGGCGCCACAAAGGUGGGAGCCACCAUUGAUCAAAUCGAUGCUACGGAAGCUCUCCAACAUCCAGGAGUGGUGGCCUUUUACACUGCGAA ..(((((..(((((....)..(((((((((((...)))))))))))))))....(((((((...)))))))....)))))...(((........)))(..(((........)))..)... ( -40.50) >DroSec_CAF1 124097 120 - 1 CACUUCCAACUCCGUUUACUGCGCCUUUGUGGGCGCCACGAAAGUGGGAGCCACCAUCGAUCAAAUCGAUGCUUCGGAAGCUCUUCAACAUCCUGGAGUGGUGGCCUUCUACACUGCGAA ..(((((..((((.......((((((....))))))(((....)))))))....(((((((...)))))))....)))))...((((......))))(..(((........)))..)... ( -42.70) >DroSim_CAF1 115101 120 - 1 CACUUCCAACUCCGUUUACUGCGCCUUCGUGGGUGCCACGAAAGUGGGAGCCACCAUCGAUCAAAUCGAUGCUUCGGAAGCUCUCCAACAUCCAGGAGUGGUGGCCUUCUACACUGCGAA ..(((((..((((.......((((((....))))))(((....)))))))....(((((((...)))))))....)))))...(((........)))(..(((........)))..)... ( -42.40) >DroEre_CAF1 111652 120 - 1 CACUUCCAACUCCGUUUACUGCGCCUUUGUGGGCGCCACAAAGGUGGGAGCCACCGUCGAUCAAAUCGAUGCUUCGGCAGCUCUCCAAUAUCCAGGAGUGGUGGCCUUUUACACUGCAAA ...................(((((((((((((...))))))))))(((.((((((((((((...))))))((....))....((((........)))).))))))))).......))).. ( -42.80) >DroYak_CAF1 114194 120 - 1 CACUUCCAACUCCGUCUACUGCGCCUUUGUGGGCGCCACAAAGGUGGGAGCCACCAUCGACCAAAUCGAUGCCUCGGAAGCUCUACAACAUCCAGGCGUGGUGGCCUUUUACACUGCGAA ............(((......(((((((((((...)))))))))))((.(((((((((((.....)))))((((.(((.(........).)))))))..))))))))........))).. ( -43.30) >consensus CACUUCCAACUCCGUUUACUGCGCCUUUGUGGGCGCCACAAAGGUGGGAGCCACCAUCGAUCAAAUCGAUGCUUCGGAAGCUCUCCAACAUCCAGGAGUGGUGGCCUUUUACACUGCGAA ..(((((..((((.......((((((....))))))(((....)))))))....((((((.....))))))....))))).................(..(((........)))..)... (-38.54 = -38.26 + -0.28)



| Location | 11,376,498 – 11,376,618 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.67 |
| Mean single sequence MFE | -41.76 |
| Consensus MFE | -37.72 |
| Energy contribution | -38.12 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.22 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.693120 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11376498 120 - 27905053 AGUGCUCCGGAGAGGCCACAUAUAUGAACGAUGUGCUGACCACUUCCAACUCCGUUUACUGCGCCUUUGUGGGCGCCACAAAGGUGGGAGCCACCAUUGAUCAAAUCGAUGCUACGGAAG .(((((((((((.((.(((((((.......))))).)).)).))))...............(((((((((((...))))))))))))))).)))(((((((...)))))))...(....) ( -39.70) >DroSec_CAF1 124137 120 - 1 AGUGCUCCGGAGAGGCCACCUACAUGAACGAUGUGCUGACCACUUCCAACUCCGUUUACUGCGCCUUUGUGGGCGCCACGAAAGUGGGAGCCACCAUCGAUCAAAUCGAUGCUUCGGAAG .....(((((((.(((..(((((.((((((..((...((.....))..))..))))))..((((((....)))))).......))))).)))..(((((((...)))))))))))))).. ( -44.20) >DroSim_CAF1 115141 120 - 1 AGUGCUCCGGAGAGGCCACCUACAUGAACGAUGUGCUGACCACUUCCAACUCCGUUUACUGCGCCUUCGUGGGUGCCACGAAAGUGGGAGCCACCAUCGAUCAAAUCGAUGCUUCGGAAG .....(((((((.((((((((((......((.(((.....))).))......(((.....))).....)))))))((((....))))..)))..(((((((...)))))))))))))).. ( -43.40) >DroEre_CAF1 111692 120 - 1 AGUGCUCCGGCGAGGCCACCUACAUGAAUGAUGUGCUGACCACUUCCAACUCCGUUUACUGCGCCUUUGUGGGCGCCACAAAGGUGGGAGCCACCGUCGAUCAAAUCGAUGCUUCGGCAG .((((((((((...))).......((((((..((...((.....))..))..))))))...(((((((((((...))))))))))))))).))).((((((...))))))((....)).. ( -40.60) >DroYak_CAF1 114234 120 - 1 AGUGCUCCGGAGAGGCCACCUACAUGAACGAUGUGCUGACCACUUCCAACUCCGUCUACUGCGCCUUUGUGGGCGCCACAAAGGUGGGAGCCACCAUCGACCAAAUCGAUGCCUCGGAAG .((((((((((((((...)))........((.(((.....))).))...))))........(((((((((((...))))))))))))))).)))((((((.....))))))......... ( -40.90) >consensus AGUGCUCCGGAGAGGCCACCUACAUGAACGAUGUGCUGACCACUUCCAACUCCGUUUACUGCGCCUUUGUGGGCGCCACAAAGGUGGGAGCCACCAUCGAUCAAAUCGAUGCUUCGGAAG .....(((((((.(((..(((((.((((((..((...((.....))..))..))))))..((((((....)))))).......))))).)))..((((((.....))))))))))))).. (-37.72 = -38.12 + 0.40)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:23:08 2006