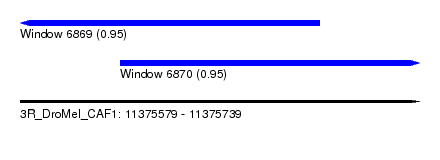
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 11,375,579 – 11,375,739 |
| Length | 160 |
| Max. P | 0.953588 |

| Location | 11,375,579 – 11,375,699 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.33 |
| Mean single sequence MFE | -37.53 |
| Consensus MFE | -35.28 |
| Energy contribution | -35.16 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.94 |
| SVM decision value | 1.37 |
| SVM RNA-class probability | 0.947104 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11375579 120 - 27905053 UACGAGGACGCUGGCUGGAACACGAACGAGAGUCCUGUGCAAGGACACUCCACUUUCACAGCAGCCAACUGCUAUGAAUUCACCGAUAGCAACUUCAAGCUGAGUGGUCAUGCUGUUCUU ...((((((((((((............(((.(((((.....))))).)))((((((((.(((((....))))).))))........((((........)))))))))))).)).)))))) ( -37.80) >DroSec_CAF1 123217 120 - 1 UACGAGGACGCUGGCUGGAACACUAACGAGAGUCCUGUGCAAGGACACUCCACUUUCACAGCAGCCAACUGCUAUGAAUUCACCGAUAGCAACUUCAAGCUGAGUGGUCAUGCUGUGCUU ...((((..((((..(((.........(((.(((((.....))))).)))....((((.(((((....))))).))))....))).))))..))))((((..(((......)))..)))) ( -37.40) >DroSim_CAF1 114221 120 - 1 UAUGAGGACGCUGGCUGGAACACGAAUGAGAGUCCUGUGCAAGGACACUCCACUUUCACAGCUGCCAACUGCUAUGAAUUCACCGAUAGCAACUUCAAGCUAAGUGGUCAUGCUGUUCUU ...((((((((((((............(((.(((((.....))))).)))(((((....((((......((((((..........))))))......))))))))))))).)).)))))) ( -34.90) >DroEre_CAF1 110774 120 - 1 UACGAGGACGCUGGCUGGAACGCGAACGAGAGUCCUGUGCAAGGACACUCCACGUUCACAGCAGCCAACUGUUAUGAAUUCACCGAUAGUAACUUCAAGCUGAGUGGUCAUGCUGUUCUU ...((((((((((.((((((((.(...(((.(((((.....))))).)))).))))).)))))))..((((((.((....))..)))))).......((((((....))).))))))))) ( -39.20) >DroYak_CAF1 113316 120 - 1 UACGAGGACGCUGGCUGGAACACGAACGAGAGUCCUGUGCAAGGACACUCCACGUUCACAGCAGCCAACUGCUACGAAUUCACCGAUAGUAACUUCAAGCUGAGUGGCCAUGCAGUUCUU ...(((((((((((((.......(((((((.(((((.....))))).)))...)))).((((.......(((((((.......)).))))).......))))...))))).)).)))))) ( -38.34) >consensus UACGAGGACGCUGGCUGGAACACGAACGAGAGUCCUGUGCAAGGACACUCCACUUUCACAGCAGCCAACUGCUAUGAAUUCACCGAUAGCAACUUCAAGCUGAGUGGUCAUGCUGUUCUU ...(((((((((((((((.........(((.(((((.....))))).)))....((((.(((((....))))).))))....))..((((........))))...))))).)).)))))) (-35.28 = -35.16 + -0.12)



| Location | 11,375,619 – 11,375,739 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.92 |
| Mean single sequence MFE | -43.26 |
| Consensus MFE | -37.90 |
| Energy contribution | -38.74 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.43 |
| SVM RNA-class probability | 0.953588 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11375619 120 + 27905053 AAUUCAUAGCAGUUGGCUGCUGUGAAAGUGGAGUGUCCUUGCACAGGACUCUCGUUCGUGUUCCAGCCAGCGUCCUCGUAAAAAUCGUUGGACAGCCCGACGAUCCUGCCACUGGUCUUC ..((((((((((....))))))))))(((((.((((....))))(((((.((.(((.(....).))).)).))))).......((((((((.....))))))))....)))))....... ( -43.00) >DroSec_CAF1 123257 120 + 1 AAUUCAUAGCAGUUGGCUGCUGUGAAAGUGGAGUGUCCUUGCACAGGACUCUCGUUAGUGUUCCAGCCAGCGUCCUCGUAGAAGUCGUUGGACAGCCCGACGAUCCUGCCACUGGUCUUC ..((((((((((....))))))))))(((((((.(((((.....))))).)))(((.((......)).)))......((((..((((((((.....)))))))).))))))))....... ( -45.00) >DroSim_CAF1 114261 120 + 1 AAUUCAUAGCAGUUGGCAGCUGUGAAAGUGGAGUGUCCUUGCACAGGACUCUCAUUCGUGUUCCAGCCAGCGUCCUCAUAGAAGUCGUUGGACAGCCCGACGAUCCUGCCACUGGUCUUC .........((((.(((((((((((.....(((.(((((.....))))).)))...(((..........)))...))))))..((((((((.....)))))))).)))))))))...... ( -40.40) >DroEre_CAF1 110814 120 + 1 AAUUCAUAACAGUUGGCUGCUGUGAACGUGGAGUGUCCUUGCACAGGACUCUCGUUCGCGUUCCAGCCAGCGUCCUCGUAGAAGUCGUUGGACAGCCCGACGAUCCUGCCACUGGUCUUC ...........((((((((....((((((((((.(((((.....))))).)))...)))))))))))))))(.((..((((..((((((((.....)))))))).))))....)).)... ( -48.30) >DroYak_CAF1 113356 120 + 1 AAUUCGUAGCAGUUGGCUGCUGUGAACGUGGAGUGUCCUUGCACAGGACUCUCGUUCGUGUUCCAGCCAGCGUCCUCGUAGAAUUCGUUGGACAGCCCGACAAUCCUGCCACUGGUCUUC ...........((((((((....((((((((((.(((((.....))))).)))...)))))))))))))))(.((..((((.(((.(((((.....)))))))).))))....)).)... ( -39.60) >consensus AAUUCAUAGCAGUUGGCUGCUGUGAAAGUGGAGUGUCCUUGCACAGGACUCUCGUUCGUGUUCCAGCCAGCGUCCUCGUAGAAGUCGUUGGACAGCCCGACGAUCCUGCCACUGGUCUUC ..((((((((((....))))))))))....(((.(((((.....))))).)))............(((((.(.....((((...(((((((.....)))))))..))))).))))).... (-37.90 = -38.74 + 0.84)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:23:03 2006