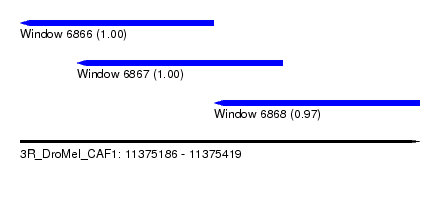
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 11,375,186 – 11,375,419 |
| Length | 233 |
| Max. P | 0.999129 |

| Location | 11,375,186 – 11,375,299 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.46 |
| Mean single sequence MFE | -41.88 |
| Consensus MFE | -38.16 |
| Energy contribution | -38.92 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.71 |
| Structure conservation index | 0.91 |
| SVM decision value | 3.39 |
| SVM RNA-class probability | 0.999129 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11375186 113 - 27905053 UCUACAUUGGGCAGUUUCCGGCAACGGUGACUAUUUACCAUGUAGAUGGCACUGUUGUGGUUACCCAUGGGGGCAUAGAAAUGGGUCAGGGUAAGUAAGGGAGCAGACUU-------UUA .............((((((.(((((((((.((((((((...)))))))))))))))))(.((((((.....(((.((....)).))).)))))).)..))))))......-------... ( -38.30) >DroSec_CAF1 122824 113 - 1 UCUACAUUGGGCAGUUUCCGGCAACAGUGACUAUUUACCAUGUCGAUGGCACUGUUGUGGUUACCCACGGGGGCAUAGAAAUGGGUCAGGGUGAGUAAGGGGACUGUCUU-------UCA ........(((((((..((.(((((((((.(((((.((...)).))))))))))))))(.((((((.....(((.((....)).))).)))))).)..))..))))))).-------... ( -43.50) >DroSim_CAF1 113828 113 - 1 UCUACAUUGGGCAGUUUCCGGCAACGGUGACUAUUUACCAUGUGGAUGGCACUGUUGUGGUUACCCAUGGGGGCAUAGAAAUGGGUCAGGGUGAGUAAGGGAGCUGUCUU-------UCA ........(((((((((((.(((((((((.((((((((...)))))))))))))))))(.((((((.....(((.((....)).))).)))))).)..))))))))))).-------... ( -46.20) >DroEre_CAF1 110382 112 - 1 UCUACAUCGGACAGUUUCCAGCAACGGUGACUAUUUACCAUGUGGAUGGCUCUGUUGUGGUUACUCAUGGGGGCAUUGAAAUGGGCCAGGGUGAGUAAG----CAGCUCUCCA----AUA ........(((.((((....(((((((.(.((((((((...))))))))).)))))))(.((((((((...(((..........)))...)))))))).----))))).))).----... ( -36.70) >DroYak_CAF1 112916 120 - 1 UCUACAUAGGGCAGUUUCCGGCAACGGUGACUAUCUACCACGUGGAUGGCACUGUUGUGGUUACCCAUGGGGGCAUAGAAAUGGGUCAGGGUGAGUAAGGGAGCUACUUACCAUAGUAUA ..(((....(((.((((((.(((((((((.((((((((...))))))))))))))))).)...(((....)))....)))))..)))..((((((((.(....)))))))))...))).. ( -44.70) >consensus UCUACAUUGGGCAGUUUCCGGCAACGGUGACUAUUUACCAUGUGGAUGGCACUGUUGUGGUUACCCAUGGGGGCAUAGAAAUGGGUCAGGGUGAGUAAGGGAGCUGCCUU_______UUA ........(((((((((((.(((((((((.((((((((...)))))))))))))))))(.((((((.....(((.((....)).))).)))))).)..)))))))))))........... (-38.16 = -38.92 + 0.76)

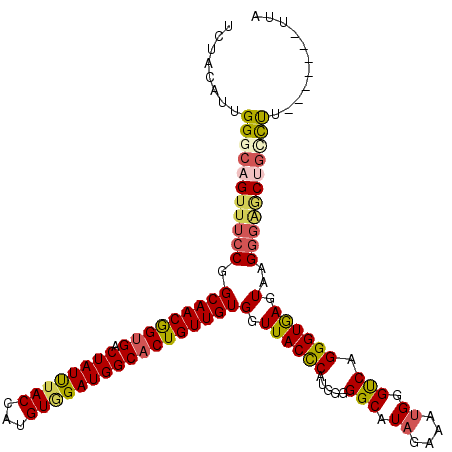
| Location | 11,375,219 – 11,375,339 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.83 |
| Mean single sequence MFE | -41.30 |
| Consensus MFE | -38.72 |
| Energy contribution | -37.24 |
| Covariance contribution | -1.48 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.94 |
| SVM decision value | 2.95 |
| SVM RNA-class probability | 0.997880 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11375219 120 - 27905053 AAAAGAGGCUUGGGUCUCUCCUUGAUGAACUUCCCCAUUAUCUACAUUGGGCAGUUUCCGGCAACGGUGACUAUUUACCAUGUAGAUGGCACUGUUGUGGUUACCCAUGGGGGCAUAGAA ...((((((....))))))............((((((((((((((((.((..((((.(((....))).)))).....))))))))))).(((....))).......)))))))....... ( -40.60) >DroSec_CAF1 122857 120 - 1 AAAAGAGGCUUGGGUCUCUCCUUGAUGAACUUCCCCGUCAUCUACAUUGGGCAGUUUCCGGCAACAGUGACUAUUUACCAUGUCGAUGGCACUGUUGUGGUUACCCACGGGGGCAUAGAA ...((((((....))))))(((.(((((.(......))))))......)))..((..((((((((((((.(((((.((...)).))))))))))))))((....)).)))..))...... ( -38.90) >DroSim_CAF1 113861 120 - 1 AAAAGAGGUUUGGGUCUCUCUUUGAUGAACUUCCCCGUCAUCUACAUUGGGCAGUUUCCGGCAACGGUGACUAUUUACCAUGUGGAUGGCACUGUUGUGGUUACCCAUGGGGGCAUAGAA .(((((((........)))))))........((((((((((((((((.((..((((.(((....))).)))).....)))))))))))))......((((....)))))))))....... ( -44.30) >DroEre_CAF1 110414 120 - 1 AAAAGGGGCUUGGGCCUCUCCUUGAUGGACUUCCCCAUCAUCUACAUCGGACAGUUUCCAGCAACGGUGACUAUUUACCAUGUGGAUGGCUCUGUUGUGGUUACUCAUGGGGGCAUUGAA ...((((((....))))))..(..(((....(((((((..(((.....))).(((..(((.((((((.(.((((((((...))))))))).)))))))))..))).))))))))))..). ( -40.10) >DroYak_CAF1 112956 120 - 1 AAAAGGGGCUUGGGUCUCUCCUUGAUGAACUUUCCCGUCAUCUACAUAGGGCAGUUUCCGGCAACGGUGACUAUCUACCACGUGGAUGGCACUGUUGUGGUUACCCAUGGGGGCAUAGAA ...((((((....))))))(((.(((((.(......)))))).....)))...((..((((((((((((.((((((((...)))))))))))))))))((....)).)))..))...... ( -42.60) >consensus AAAAGAGGCUUGGGUCUCUCCUUGAUGAACUUCCCCGUCAUCUACAUUGGGCAGUUUCCGGCAACGGUGACUAUUUACCAUGUGGAUGGCACUGUUGUGGUUACCCAUGGGGGCAUAGAA ...((((((....))))))...(((((........))))).............((..((((((((((((.((((((((...)))))))))))))))))((....)).)))..))...... (-38.72 = -37.24 + -1.48)

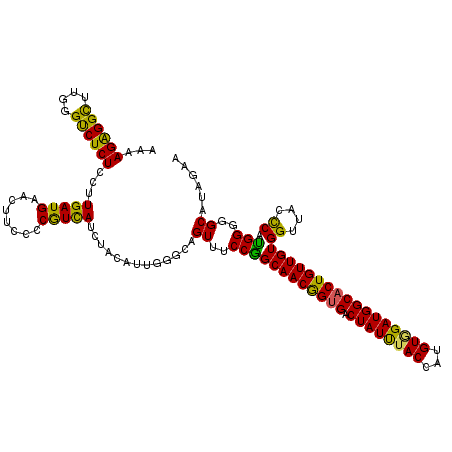
| Location | 11,375,299 – 11,375,419 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.58 |
| Mean single sequence MFE | -37.87 |
| Consensus MFE | -32.81 |
| Energy contribution | -33.33 |
| Covariance contribution | 0.52 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.66 |
| SVM RNA-class probability | 0.970671 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11375299 120 - 27905053 UUCCAGAGUUCCUCAAGACUAGGGAGUACUACUCCCGGAAGAAGGAGAUCGAGGCAUUCAAUGCCAACAAUCGAUGGAAAAAAAGAGGCUUGGGUCUCUCCUUGAUGAACUUCCCCAUUA .....((((((.(((((....(((((.....))))).......(((((((.((((.(((....(((.(....).))).......))))))).))))))).))))).))))))........ ( -38.70) >DroSec_CAF1 122937 120 - 1 UUCCAGAGUUCCUCAAGACUAGGGAGUACUACUCCCGGAAGAAGGAGAUCGAGAAAUUCAAUGCCAACAAUCGAUGGAAAAAAAGAGGCUUGGGUCUCUCCUUGAUGAACUUCCCCGUCA .....(((...)))..(((..(((((.....)))))((..((((...((((((..........(((.(....).)))......((((((....)))))).))))))...)))).))))). ( -33.50) >DroSim_CAF1 113941 120 - 1 UUCCAGAGUUCCUCAAGACUAGGGAGUACUACUCCCGGAAGAAGGAGAUCGAGGCAUUCAAUGCCAACAACCGAUGGAAAAAAAGAGGUUUGGGUCUCUCUUUGAUGAACUUCCCCGUCA ((((..((((......)))).(((((.....)))))))))((((((((((..(((((...)))))..(((((..(........)..)).))))))))).))))((((........)))). ( -36.70) >DroEre_CAF1 110494 120 - 1 UUCCGGAGUUCCUCAAGACUAGGGAGUACUACUCCCGGAGGAAGGAGAUCGAGCAGUUCAAUGCCAACAACCGGUGGAAGAAAAGGGGCUUGGGCCUCUCCUUGAUGGACUUCCCCAUCA ..((((..((((((.......(((((.....))))).)))))).......(.(((......)))).....)))).(((((....(((((....)))))(((.....))))))))...... ( -41.90) >DroYak_CAF1 113036 120 - 1 UUCCGGAGUUCCUCAAGACUAGGGAGUACUACUCCCGGCGGAAGGAGAUCGAGGUAUUCAACGCCAACAACCGAUGGAAAAAAAGGGGCUUGGGUCUCUCCUUGAUGAACUUUCCCGUCA ...((((((((.(((((....(((((.....))))).......(((((((.((((........(((.(....).)))..........)))).))))))).))))).)))))...)))... ( -38.57) >consensus UUCCAGAGUUCCUCAAGACUAGGGAGUACUACUCCCGGAAGAAGGAGAUCGAGGAAUUCAAUGCCAACAACCGAUGGAAAAAAAGAGGCUUGGGUCUCUCCUUGAUGAACUUCCCCGUCA .....((((((.(((((....(((((.....))))).......(((((((.((((........(((.(....).)))..........)))).))))))).))))).))))))........ (-32.81 = -33.33 + 0.52)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:23:01 2006