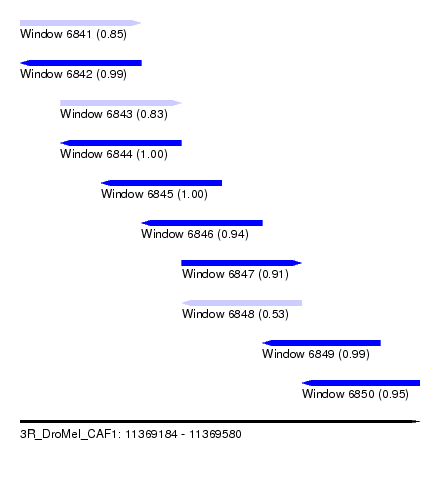
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 11,369,184 – 11,369,580 |
| Length | 396 |
| Max. P | 0.999799 |

| Location | 11,369,184 – 11,369,304 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.33 |
| Mean single sequence MFE | -33.04 |
| Consensus MFE | -28.84 |
| Energy contribution | -28.88 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.845483 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11369184 120 + 27905053 UGGCAUCCUUCUUCUUCACCGGCUCCAAGCGACUGUUCAAGGUCUCACAGGCCUUCCGAACUGCAUAGCAGAGACUUUCGCUGCCCACAGCUCCUCCGGUGACCAUCGAAUUGGCUCCAU (((............(((((((.....(((((..(((.((((((.....)))))).....((((...)))).)))..)))))((.....))....)))))))(((......)))..))). ( -33.70) >DroSec_CAF1 116820 120 + 1 UGGCGUCCUUCUUCUUCACCGGCUCCAAACGACUGUUCAAGGUCUCACAGGCCUUUCGAACUGCAUAGCAGAGACUUUCGCUGCCCACUGCUCCUCCGGUGACCAUCGAAUUGGCUCCAU (((.(((.(((....(((((((............((((((((((.....))))))..)))).(((..((((.(.....).))))....)))....))))))).....)))..))).))). ( -34.80) >DroSim_CAF1 107849 120 + 1 UGGCAUCCUUCUUCUUCACCGGCUCCAAACGACUGUUCAAGGUCUCACAGGCCUUUCGAACUGCAUAGCAGAGACUUUCGCUGCCCACAGCUCCUCCGGUGACCAUCGAAUUGGCUCCAU (((............((((((((((.........((((((((((.....))))))..))))(((...))))))......((((....))))....)))))))(((......)))..))). ( -32.00) >DroEre_CAF1 104603 120 + 1 UGGCAUCCUUCUUCUUCACCGGCUCCAAGCGACUGUUUAAGGUCUCACAGGCCUUGCGAACUGCAUAGCAGAGACUUUCACUGCCAACAGCUCCUCCGGUGACCAUCGAAUUGGCUCCAU (((............(((((((.....((.((((((((((((((.....)))))))...........((((.(.....).)))).))))).)))))))))))(((......)))..))). ( -32.10) >DroYak_CAF1 106929 120 + 1 UGGCAUCCUUCUUCCUCACCGGCUCCAAACGACUGUUCAAAGUCUCGCAGGCCUUGCGAACUGCAAAGCAGAGACUUUCACUGCCAACUGCUCCUCCGGUGACCAUCGAAUUGGCUCCAU (((............(((((((........((((......))))..((((...((((.....)))).((((.(.....).))))...))))....)))))))(((......)))..))). ( -32.60) >consensus UGGCAUCCUUCUUCUUCACCGGCUCCAAACGACUGUUCAAGGUCUCACAGGCCUUGCGAACUGCAUAGCAGAGACUUUCGCUGCCCACAGCUCCUCCGGUGACCAUCGAAUUGGCUCCAU (((............(((((((............((((((((((.....))))))..)))).((...((((.(.....).)))).....))....)))))))(((......)))..))). (-28.84 = -28.88 + 0.04)


| Location | 11,369,184 – 11,369,304 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.33 |
| Mean single sequence MFE | -42.68 |
| Consensus MFE | -39.82 |
| Energy contribution | -39.42 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.93 |
| SVM decision value | 2.11 |
| SVM RNA-class probability | 0.988305 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11369184 120 - 27905053 AUGGAGCCAAUUCGAUGGUCACCGGAGGAGCUGUGGGCAGCGAAAGUCUCUGCUAUGCAGUUCGGAAGGCCUGUGAGACCUUGAACAGUCGCUUGGAGCCGGUGAAGAAGAAGGAUGCCA .(((..((..(((.....(((((((..(((((((((((((.(.....).))))).))))))))...((((((((..........))))..))))....))))))).)))...))...))) ( -41.30) >DroSec_CAF1 116820 120 - 1 AUGGAGCCAAUUCGAUGGUCACCGGAGGAGCAGUGGGCAGCGAAAGUCUCUGCUAUGCAGUUCGAAAGGCCUGUGAGACCUUGAACAGUCGUUUGGAGCCGGUGAAGAAGAAGGACGCCA .(((..((..(((.....(((((((.(((((((.((((.......)))))))))..((((..(....)..))))....))(..(((....)))..)..))))))).)))...))...))) ( -41.60) >DroSim_CAF1 107849 120 - 1 AUGGAGCCAAUUCGAUGGUCACCGGAGGAGCUGUGGGCAGCGAAAGUCUCUGCUAUGCAGUUCGAAAGGCCUGUGAGACCUUGAACAGUCGUUUGGAGCCGGUGAAGAAGAAGGAUGCCA .(((..((..(((.....(((((((....((((....))))....(((((......((((..(....)..))))))))).(..(((....)))..)..))))))).)))...))...))) ( -40.70) >DroEre_CAF1 104603 120 - 1 AUGGAGCCAAUUCGAUGGUCACCGGAGGAGCUGUUGGCAGUGAAAGUCUCUGCUAUGCAGUUCGCAAGGCCUGUGAGACCUUAAACAGUCGCUUGGAGCCGGUGAAGAAGAAGGAUGCCA .(((..((..(((.....(((((((..(((((((((((((.(.....).)))))).))))))).(((((.((((..........)))).).))))...))))))).)))...))...))) ( -44.90) >DroYak_CAF1 106929 120 - 1 AUGGAGCCAAUUCGAUGGUCACCGGAGGAGCAGUUGGCAGUGAAAGUCUCUGCUUUGCAGUUCGCAAGGCCUGCGAGACUUUGAACAGUCGUUUGGAGCCGGUGAGGAAGAAGGAUGCCA .(((..((..(((.....(((((((.(((((((..(((.......))).)))))))((((..(....)..))))....(((..(((....)))..))))))))))....)))))...))) ( -44.90) >consensus AUGGAGCCAAUUCGAUGGUCACCGGAGGAGCUGUGGGCAGCGAAAGUCUCUGCUAUGCAGUUCGAAAGGCCUGUGAGACCUUGAACAGUCGUUUGGAGCCGGUGAAGAAGAAGGAUGCCA .(((..((..(((.....(((((((..((((....(((((.(.....).)))))..((((..(....)..))))..(((........)))))))....))))))).)))...))...))) (-39.82 = -39.42 + -0.40)


| Location | 11,369,224 – 11,369,344 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.17 |
| Mean single sequence MFE | -38.34 |
| Consensus MFE | -35.70 |
| Energy contribution | -35.42 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.29 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.825713 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11369224 120 + 27905053 GGUCUCACAGGCCUUCCGAACUGCAUAGCAGAGACUUUCGCUGCCCACAGCUCCUCCGGUGACCAUCGAAUUGGCUCCAUUGAUGGUGUCGGAGGACUCCACCUUGAUGAAGCUCAAAUC ((((.....)))).......((((...)))).((.((((((((....))))(((((((..(.(((((((..((....))))))))))..)))))))............)))).))..... ( -40.40) >DroSec_CAF1 116860 120 + 1 GGUCUCACAGGCCUUUCGAACUGCAUAGCAGAGACUUUCGCUGCCCACUGCUCCUCCGGUGACCAUCGAAUUGGCUCCAUUGAUGGUGUCGGAGGACUCCACCUUGAUGAAGCUCAAAUC ((..(((((((...........(((..((((.(.....).))))....)))(((((((..(.(((((((..((....))))))))))..)))))))......)))).)))..))...... ( -38.00) >DroSim_CAF1 107889 120 + 1 GGUCUCACAGGCCUUUCGAACUGCAUAGCAGAGACUUUCGCUGCCCACAGCUCCUCCGGUGACCAUCGAAUUGGCUCCAUUGAUAGUGUCGGAGGACUCCACCUUGAUGAAGCUCAAAUC ((((.....)))).......((((...)))).((.((((((((....))))(((((((..(.(.(((((..((....))))))).))..)))))))............)))).))..... ( -33.90) >DroEre_CAF1 104643 120 + 1 GGUCUCACAGGCCUUGCGAACUGCAUAGCAGAGACUUUCACUGCCAACAGCUCCUCCGGUGACCAUCGAAUUGGCUCCAUUGAUGGUGUCGGAGGACUCCACCUUGAUGAAGCCCAAAUC ((..(((((((...((.((.(((....((((.(.....).))))...))).(((((((..(.(((((((..((....))))))))))..))))))).)))).)))).)))..))...... ( -39.60) >DroYak_CAF1 106969 120 + 1 AGUCUCGCAGGCCUUGCGAACUGCAAAGCAGAGACUUUCACUGCCAACUGCUCCUCCGGUGACCAUCGAAUUGGCUCCAUUGAUGGUGUCGGAGGACUCCACCUUGAUGAAGCUCAAAUC ((..((((((...((((.....)))).((((.(.....).))))...))))(((((((..(.(((((((..((....))))))))))..)))))))............))..))...... ( -39.80) >consensus GGUCUCACAGGCCUUGCGAACUGCAUAGCAGAGACUUUCGCUGCCCACAGCUCCUCCGGUGACCAUCGAAUUGGCUCCAUUGAUGGUGUCGGAGGACUCCACCUUGAUGAAGCUCAAAUC ((..(((((((...........((...((((.(.....).)))).....))(((((((..(.(((((((..((....))))))))))..)))))))......)))).)))..))...... (-35.70 = -35.42 + -0.28)



| Location | 11,369,224 – 11,369,344 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.17 |
| Mean single sequence MFE | -45.50 |
| Consensus MFE | -44.28 |
| Energy contribution | -43.96 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.97 |
| SVM decision value | 3.57 |
| SVM RNA-class probability | 0.999404 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11369224 120 - 27905053 GAUUUGAGCUUCAUCAAGGUGGAGUCCUCCGACACCAUCAAUGGAGCCAAUUCGAUGGUCACCGGAGGAGCUGUGGGCAGCGAAAGUCUCUGCUAUGCAGUUCGGAAGGCCUGUGAGACC ((.(((.(((((((...(((((.(((....))).))))).)))))))))).))...((((..((((((.((((....)))).....))))))....((((..(....)..))))..)))) ( -46.60) >DroSec_CAF1 116860 120 - 1 GAUUUGAGCUUCAUCAAGGUGGAGUCCUCCGACACCAUCAAUGGAGCCAAUUCGAUGGUCACCGGAGGAGCAGUGGGCAGCGAAAGUCUCUGCUAUGCAGUUCGAAAGGCCUGUGAGACC ((.(((.(((((((...(((((.(((....))).))))).)))))))))).))...((((........(((((.((((.......)))))))))..((((..(....)..))))..)))) ( -44.40) >DroSim_CAF1 107889 120 - 1 GAUUUGAGCUUCAUCAAGGUGGAGUCCUCCGACACUAUCAAUGGAGCCAAUUCGAUGGUCACCGGAGGAGCUGUGGGCAGCGAAAGUCUCUGCUAUGCAGUUCGAAAGGCCUGUGAGACC ((.(((.(((((((...(((((.(((....))).))))).)))))))))).))...((((..((((((.((((....)))).....))))))....((((..(....)..))))..)))) ( -44.30) >DroEre_CAF1 104643 120 - 1 GAUUUGGGCUUCAUCAAGGUGGAGUCCUCCGACACCAUCAAUGGAGCCAAUUCGAUGGUCACCGGAGGAGCUGUUGGCAGUGAAAGUCUCUGCUAUGCAGUUCGCAAGGCCUGUGAGACC ((....((((((((...(((((.(((....))).))))).))))))))...))...((((..((.(((((((((((((((.(.....).)))))).))))))(....).))).)).)))) ( -46.40) >DroYak_CAF1 106969 120 - 1 GAUUUGAGCUUCAUCAAGGUGGAGUCCUCCGACACCAUCAAUGGAGCCAAUUCGAUGGUCACCGGAGGAGCAGUUGGCAGUGAAAGUCUCUGCUUUGCAGUUCGCAAGGCCUGCGAGACU ((.(((.(((((((...(((((.(((....))).))))).)))))))))).))...((((......(((((((..(((.......))).)))))))((((..(....)..))))..)))) ( -45.80) >consensus GAUUUGAGCUUCAUCAAGGUGGAGUCCUCCGACACCAUCAAUGGAGCCAAUUCGAUGGUCACCGGAGGAGCUGUGGGCAGCGAAAGUCUCUGCUAUGCAGUUCGAAAGGCCUGUGAGACC ((.(((.(((((((...(((((.(((....))).))))).)))))))))).))...((((..((((((.((((....)))).....))))))....((((..(....)..))))..)))) (-44.28 = -43.96 + -0.32)


| Location | 11,369,264 – 11,369,384 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.00 |
| Mean single sequence MFE | -50.02 |
| Consensus MFE | -46.34 |
| Energy contribution | -47.06 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.57 |
| Structure conservation index | 0.93 |
| SVM decision value | 4.11 |
| SVM RNA-class probability | 0.999799 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11369264 120 - 27905053 CACUAAGGUGGCUCAAGUGGCUGCCUACACCCUUGGCAUCGAUUUGAGCUUCAUCAAGGUGGAGUCCUCCGACACCAUCAAUGGAGCCAAUUCGAUGGUCACCGGAGGAGCUGUGGGCAG ((((.((....))..)))).((((((((((((((((((((((.(((.(((((((...(((((.(((....))).))))).)))))))))).))))))....)).)))).).))))))))) ( -50.00) >DroSec_CAF1 116900 120 - 1 UACUAAGGUGGCACAGGUGGCUGCCUACACCCUGGGCAUCGAUUUGAGCUUCAUCAAGGUGGAGUCCUCCGACACCAUCAAUGGAGCCAAUUCGAUGGUCACCGGAGGAGCAGUGGGCAG ..((..((((((.((((((........)).))))..((((((.(((.(((((((...(((((.(((....))).))))).)))))))))).))))))))))))..))..((.....)).. ( -49.40) >DroSim_CAF1 107929 120 - 1 CACUAAGGUGGCACAGGUGGCUGCCUACACCCUGGGCAUCGAUUUGAGCUUCAUCAAGGUGGAGUCCUCCGACACUAUCAAUGGAGCCAAUUCGAUGGUCACCGGAGGAGCUGUGGGCAG (((....)))..........(((((((((((((((.((((((.(((.(((((((...(((((.(((....))).))))).)))))))))).))))))....)))).))...))))))))) ( -49.30) >DroEre_CAF1 104683 120 - 1 CACUAAGGUCGCUCAGGUGGCUGCCUACACCCUGGGCAUCGAUUUGGGCUUCAUCAAGGUGGAGUCCUCCGACACCAUCAAUGGAGCCAAUUCGAUGGUCACCGGAGGAGCUGUUGGCAG ((...((.((.(((.((((((((((((.....)))))(((((....((((((((...(((((.(((....))).))))).))))))))...)))))))))))).))))).))..)).... ( -51.90) >DroYak_CAF1 107009 120 - 1 CACUAAGGUGGCGCAGGUGGCUGCCUUCACCCUGGGCAUCGAUUUGAGCUUCAUCAAGGUGGAGUCCUCCGACACCAUCAAUGGAGCCAAUUCGAUGGUCACCGGAGGAGCAGUUGGCAG (((....))).....(.(((((((((((....(((.((((((.(((.(((((((...(((((.(((....))).))))).)))))))))).)))))).)))...)))).))))))).).. ( -49.50) >consensus CACUAAGGUGGCACAGGUGGCUGCCUACACCCUGGGCAUCGAUUUGAGCUUCAUCAAGGUGGAGUCCUCCGACACCAUCAAUGGAGCCAAUUCGAUGGUCACCGGAGGAGCUGUGGGCAG ((((...........)))).(((((((((((((((.((((((.(((.(((((((...(((((.(((....))).))))).)))))))))).))))))....)))).))...))))))))) (-46.34 = -47.06 + 0.72)



| Location | 11,369,304 – 11,369,424 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.17 |
| Mean single sequence MFE | -33.46 |
| Consensus MFE | -30.10 |
| Energy contribution | -30.22 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.28 |
| SVM RNA-class probability | 0.939156 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11369304 120 - 27905053 UAUUUCAUUGAUAACAUAUUGAUUUUCCUAUAGGUAUGAACACUAAGGUGGCUCAAGUGGCUGCCUACACCCUUGGCAUCGAUUUGAGCUUCAUCAAGGUGGAGUCCUCCGACACCAUCA .......(((((..(((((..((......))..)))))..(((....)))((((((((.(.((((.........)))).).))))))))...)))))(((((.(((....))).))))). ( -36.30) >DroSec_CAF1 116940 119 - 1 UAUU-UAUUGAUAACAUAUUGAUUUUCUUAUAGGUAUGAAUACUAAGGUGGCACAGGUGGCUGCCUACACCCUGGGCAUCGAUUUGAGCUUCAUCAAGGUGGAGUCCUCCGACACCAUCA ....-..(((((..(((((..((......))..))))).........(.(((.(((((.(.((((((.....)))))).).))))).))).))))))(((((.(((....))).))))). ( -32.70) >DroSim_CAF1 107969 120 - 1 UAUUUUCUUGAUAACAUAUUGAUUUUCCUAUAGGUAUGAACACUAAGGUGGCACAGGUGGCUGCCUACACCCUGGGCAUCGAUUUGAGCUUCAUCAAGGUGGAGUCCUCCGACACUAUCA .......(((((..(((((..((......))..)))))..(((....)))((.(((((.(.((((((.....)))))).).))))).))...)))))(((((.(((....))).))))). ( -30.60) >DroEre_CAF1 104723 120 - 1 CAUUUCAUUGACAACAUGUUGAUUUUCUUAUAGGUAUGAACACUAAGGUCGCUCAGGUGGCUGCCUACACCCUGGGCAUCGAUUUGGGCUUCAUCAAGGUGGAGUCCUCCGACACCAUCA .........(((....((((.((...((....)).)).)))).....)))((((((((.(.((((((.....)))))).).))))))))........(((((.(((....))).))))). ( -34.80) >DroYak_CAF1 107049 120 - 1 CAUUUCAUUGAUAACAUAUUGAUUUUCCUAUAGGUAUGAACACUAAGGUGGCGCAGGUGGCUGCCUUCACCCUGGGCAUCGAUUUGAGCUUCAUCAAGGUGGAGUCCUCCGACACCAUCA .......(((((..(((((..((......))..)))))..(((....)))((.(((((.(.(((((.......))))).).))))).))...)))))(((((.(((....))).))))). ( -32.90) >consensus UAUUUCAUUGAUAACAUAUUGAUUUUCCUAUAGGUAUGAACACUAAGGUGGCACAGGUGGCUGCCUACACCCUGGGCAUCGAUUUGAGCUUCAUCAAGGUGGAGUCCUCCGACACCAUCA .......(((((..(((((..((......))..))))).........(..((.(((((.(.((((((.....)))))).).))))).))..))))))(((((.(((....))).))))). (-30.10 = -30.22 + 0.12)


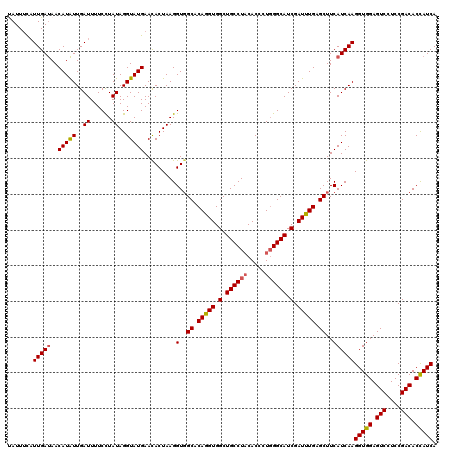
| Location | 11,369,344 – 11,369,463 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.95 |
| Mean single sequence MFE | -33.72 |
| Consensus MFE | -24.82 |
| Energy contribution | -25.98 |
| Covariance contribution | 1.16 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.58 |
| Structure conservation index | 0.74 |
| SVM decision value | 1.05 |
| SVM RNA-class probability | 0.907287 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11369344 119 + 27905053 GAUGCCAAGGGUGUAGGCAGCCACUUGAGCCACCUUAGUGUUCAUACCUAUAGGAAAAUCAAUAUGUUAUCAAUGAAAUAGUAU-CUUAAGUGGGCUGUUUACUUACCCUGUCCCAUUUC ((((...((((((((((((((((((((((...((((((.((....))))).)))....(((...((....)).)))........-))))))).))))))))))..)))))....)))).. ( -34.60) >DroSec_CAF1 116980 118 + 1 GAUGCCCAGGGUGUAGGCAGCCACCUGUGCCACCUUAGUAUUCAUACCUAUAAGAAAAUCAAUAUGUUAUCAAUA-AAUAAUAU-CUUAAGUGGGCUGUUUACUCACCCUGUCCCAUUUC ((((..((((((((((((((((..((((((.......)))).........(((((.........((((((.....-.)))))))-))))))..))))))))))..))))))...)))).. ( -29.10) >DroSim_CAF1 108009 120 + 1 GAUGCCCAGGGUGUAGGCAGCCACCUGUGCCACCUUAGUGUUCAUACCUAUAGGAAAAUCAAUAUGUUAUCAAGAAAAUAAUAAAAUUAAAGGGGCUGUUUACUUACCCUGUCCCAUUUC ((((..((((((((((((((((.(((......((((((.((....))))).)))..........((((((.......)))))).......)))))))))))))..))))))...)))).. ( -35.30) >DroEre_CAF1 104763 118 + 1 GAUGCCCAGGGUGUAGGCAGCCACCUGAGCGACCUUAGUGUUCAUACCUAUAAGAAAAUCAACAUGUUGUCAAUGAAAUGACAU-CUUAAGG-GGCUGAUUACUUACCCUGUCCCAUUUC ((((..((((((((((.(((((...(((((.((....)))))))..(((.(((((............(((((......))))))-)))))))-))))).))))..))))))...)))).. ( -34.80) >DroYak_CAF1 107089 119 + 1 GAUGCCCAGGGUGAAGGCAGCCACCUGCGCCACCUUAGUGUUCAUACCUAUAGGAAAAUCAAUAUGUUAUCAAUGAAAUGAUAU-CUUAAGGGGGCUGAUUACUCACCCUGUCCCAUUUC ((((..((((((((.(.(((((.(((......((((((.((....))))).))).............(((((......))))).-....))).)))))....)))))))))...)))).. ( -34.80) >consensus GAUGCCCAGGGUGUAGGCAGCCACCUGAGCCACCUUAGUGUUCAUACCUAUAGGAAAAUCAAUAUGUUAUCAAUGAAAUAAUAU_CUUAAGGGGGCUGUUUACUUACCCUGUCCCAUUUC ((((..((((((((((((((((.(((......((((((.((....))))).)))..........((((((.......))))))......))).))))))))))..))))))...)))).. (-24.82 = -25.98 + 1.16)

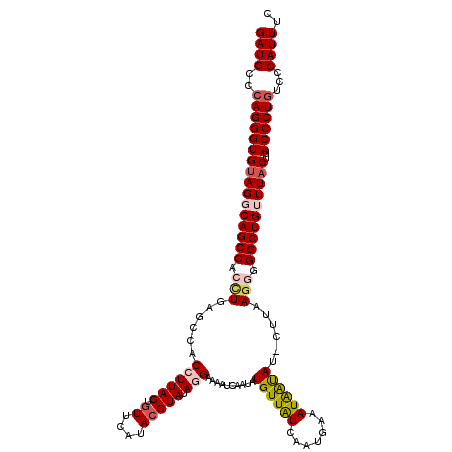

| Location | 11,369,344 – 11,369,463 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.95 |
| Mean single sequence MFE | -32.62 |
| Consensus MFE | -25.12 |
| Energy contribution | -25.56 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.77 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.528080 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11369344 119 - 27905053 GAAAUGGGACAGGGUAAGUAAACAGCCCACUUAAG-AUACUAUUUCAUUGAUAACAUAUUGAUUUUCCUAUAGGUAUGAACACUAAGGUGGCUCAAGUGGCUGCCUACACCCUUGGCAUC .........((((((..(((..(((((.((((.((-..................(((((..((......))..)))))..(((....))).)).)))))))))..))))))).))..... ( -26.60) >DroSec_CAF1 116980 118 - 1 GAAAUGGGACAGGGUGAGUAAACAGCCCACUUAAG-AUAUUAUU-UAUUGAUAACAUAUUGAUUUUCUUAUAGGUAUGAAUACUAAGGUGGCACAGGUGGCUGCCUACACCCUGGGCAUC ......(..(((((((((((..((..((...((((-(.((((..-...((....))...))))..)))))..))..))..)))).(((..((.(....)))..))).)))))))..)... ( -32.30) >DroSim_CAF1 108009 120 - 1 GAAAUGGGACAGGGUAAGUAAACAGCCCCUUUAAUUUUAUUAUUUUCUUGAUAACAUAUUGAUUUUCCUAUAGGUAUGAACACUAAGGUGGCACAGGUGGCUGCCUACACCCUGGGCAUC ......(..((((((..(((..((((((((......((((((......))))))(((((..((......))..)))))..(((....)))....))).)))))..)))))))))..)... ( -31.90) >DroEre_CAF1 104763 118 - 1 GAAAUGGGACAGGGUAAGUAAUCAGCC-CCUUAAG-AUGUCAUUUCAUUGACAACAUGUUGAUUUUCUUAUAGGUAUGAACACUAAGGUCGCUCAGGUGGCUGCCUACACCCUGGGCAUC ......(..((((((..(((..(((((-(((...(-((((((......))))....((((.((...((....)).)).)))).....)))....))).)))))..)))))))))..)... ( -33.50) >DroYak_CAF1 107089 119 - 1 GAAAUGGGACAGGGUGAGUAAUCAGCCCCCUUAAG-AUAUCAUUUCAUUGAUAACAUAUUGAUUUUCCUAUAGGUAUGAACACUAAGGUGGCGCAGGUGGCUGCCUUCACCCUGGGCAUC ......(..(((((((((......(((.(((((..-.(((((......))))).(((((..((......))..))))).....))))).)))((((....)))).)))))))))..)... ( -38.80) >consensus GAAAUGGGACAGGGUAAGUAAACAGCCCCCUUAAG_AUAUUAUUUCAUUGAUAACAUAUUGAUUUUCCUAUAGGUAUGAACACUAAGGUGGCACAGGUGGCUGCCUACACCCUGGGCAUC ......(..((((((..(((..(((((.(((......(((((......))))).(((((..((......))..)))))..(((....)))....))).)))))..)))))))))..)... (-25.12 = -25.56 + 0.44)

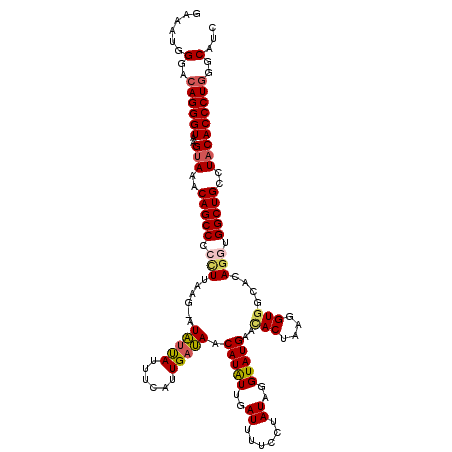

| Location | 11,369,424 – 11,369,541 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.47 |
| Mean single sequence MFE | -39.20 |
| Consensus MFE | -35.24 |
| Energy contribution | -34.96 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.90 |
| SVM decision value | 2.32 |
| SVM RNA-class probability | 0.992255 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11369424 117 - 27905053 UUCUAUUUU-GGCCAGUACC-CGGCUACAGUUGCCAUCUAUCAUGUGGAUGGCACUGUUGUGGUUACCCAUGGAGGCAUUGAAAUGGGACAGGGUAAGUAAACAGCCCACUUAAG-AUAC (((((((((-.(((.(((.(-((.(.(((((.(((((((((...)))))))))))))).)))).)))(....).)))...)))))))))..((((.........)))).......-.... ( -38.80) >DroSec_CAF1 117059 117 - 1 UUCUAUUUC-GGCCAGUACC-CGGCUACAGUGGCCAUCUAUCAUGUGGAUGGCACUGUUGUGGUUACCCACGGAGGCAUUGAAAUGGGACAGGGUGAGUAAACAGCCCACUUAAG-AUAU (((((((((-.(((......-.....((((((.((((((((...))))))))))))))(((((....)))))..)))...)))))))))..((((..(....).)))).......-.... ( -41.30) >DroSim_CAF1 108089 120 - 1 UUCUAUUUUGGGCCAGUACCCCGGCUACAGUGGCCAUCUAUCAUGUGGAUGGCACUGUUGUGGUUUCCCAUGGAGGCAUUGAAAUGGGACAGGGUAAGUAAACAGCCCCUUUAAUUUUAU .........((((...((((((.((.((((((.((((((((...)))))))))))))).)).)..((((((............))))))..)))))........))))............ ( -40.50) >DroEre_CAF1 104843 116 - 1 UUCUAUUUU-GGUCAGUACC-CGGCUACUGUGGCCAUCUAUCAUGUGGAUGGCACUGUUGUGGUUACCCAUGGAGGCAUCGAAAUGGGACAGGGUAAGUAAUCAGCC-CCUUAAG-AUGU .......((-(((...((((-(((((((.(((.((((((((...)))))))))))....)))))).(((((.((....))...)))))...)))))....)))))..-.......-.... ( -35.20) >DroYak_CAF1 107169 117 - 1 UUCUAUUUU-GGUCAGUACC-CGGCUACGGUGGCCAUCUAUCAUGUGGAUGGCACUGUUGUGGUUACCCAUGGAGGCAUCGAAAUGGGACAGGGUGAGUAAUCAGCCCCCUUAAG-AUAU (((((((((-(((..((..(-((((.((((((.((((((((...)))))))))))))).))((....)).)))..))))))))))))))..((((((....))).))).......-.... ( -40.20) >consensus UUCUAUUUU_GGCCAGUACC_CGGCUACAGUGGCCAUCUAUCAUGUGGAUGGCACUGUUGUGGUUACCCAUGGAGGCAUUGAAAUGGGACAGGGUAAGUAAACAGCCCCCUUAAG_AUAU (((((((((..(((............(((((.(((((((((...)))))))))))))).((((....))))...)))...)))))))))..((((.........))))............ (-35.24 = -34.96 + -0.28)



| Location | 11,369,463 – 11,369,580 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.30 |
| Mean single sequence MFE | -41.06 |
| Consensus MFE | -37.94 |
| Energy contribution | -37.34 |
| Covariance contribution | -0.60 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.37 |
| SVM RNA-class probability | 0.947264 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11369463 117 - 27905053 AAACGAGGUCUCGGAUUGGCGGU-AAUGGACUACCCCAUUUUCUAUUUU-GGCCAGUACC-CGGCUACAGUUGCCAUCUAUCAUGUGGAUGGCACUGUUGUGGUUACCCAUGGAGGCAUU .......(((((((((((((.(.-((((((...........)))))).)-.)))))).))-(.((.(((((.(((((((((...)))))))))))))).)).).........)))))... ( -42.30) >DroSec_CAF1 117098 117 - 1 AAGCGAGGUCUCGGAUUGGCGGU-GAUGGACUAUCCCAUUUUCUAUUUC-GGCCAGUACC-CGGCUACAGUGGCCAUCUAUCAUGUGGAUGGCACUGUUGUGGUUACCCACGGAGGCAUU .......(((((((((((((.(.-((((((...........)))))).)-.)))))).))-.....((((((.((((((((...))))))))))))))(((((....))))))))))... ( -44.70) >DroSim_CAF1 108129 120 - 1 AAGCGAGGUCUCGGAUUGGCGGCGGAUGGACAACCCCAUGUUCUAUUUUGGGCCAGUACCCCGGCUACAGUGGCCAUCUAUCAUGUGGAUGGCACUGUUGUGGUUUCCCAUGGAGGCAUU .......(((((((((((((...(((((((((......)).)))))))...)))))).)).(.((.((((((.((((((((...)))))))))))))).)).).........)))))... ( -43.00) >DroEre_CAF1 104881 117 - 1 AAACGAGGUCUUGGACUGGCGGU-UAUGGACUAUCCCAUUUUCUAUUUU-GGUCAGUACC-CGGCUACUGUGGCCAUCUAUCAUGUGGAUGGCACUGUUGUGGUUACCCAUGGAGGCAUC .......(((((((((((((.(.-.(((((...........)))))..)-.)))))).))-.((((((.(((.((((((((...)))))))))))....)))))).......)))))... ( -36.60) >DroYak_CAF1 107208 117 - 1 AAACGAGGUCUUGGACUGGCGGU-AAUGGACUAUCCCAUUUUCUAUUUU-GGUCAGUACC-CGGCUACGGUGGCCAUCUAUCAUGUGGAUGGCACUGUUGUGGUUACCCAUGGAGGCAUC .......(((((((((((((.(.-((((((...........)))))).)-.)))))).))-(.((.((((((.((((((((...)))))))))))))).)).).........)))))... ( -38.70) >consensus AAACGAGGUCUCGGAUUGGCGGU_AAUGGACUAUCCCAUUUUCUAUUUU_GGCCAGUACC_CGGCUACAGUGGCCAUCUAUCAUGUGGAUGGCACUGUUGUGGUUACCCAUGGAGGCAUU .......(((((((((((((....((((((...........))))))....)))))).)).(.((.(((((.(((((((((...)))))))))))))).)).).........)))))... (-37.94 = -37.34 + -0.60)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:22:43 2006