
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 11,367,840 – 11,368,040 |
| Length | 200 |
| Max. P | 0.999780 |

| Location | 11,367,840 – 11,367,960 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.61 |
| Mean single sequence MFE | -47.92 |
| Consensus MFE | -38.12 |
| Energy contribution | -39.44 |
| Covariance contribution | 1.32 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.630757 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11367840 120 + 27905053 UUAUUGCUCGCUGUAUUCAGGUGUGGAGAAAGCCACUCCCCGAAAAGUGACUUCUUACCGAGUUGGCUGCCCUGGACUGAACCUCCUGGGUGACCGGGUGGCGCCUGCGGAUGACGCAGA ...(((((((((...(((.((.((((......)))).))..))).))))).......(((....(((.(((..((......)).(((((....))))).))))))..))).....)))). ( -42.90) >DroSec_CAF1 115478 112 + 1 UUAUUGCUCGCUG--------UGGGGAGAAAGCCACUCCCCGAAAAGCGACUUCUUACCGAGUUGGCUGCACGGGACUGAACCUCCUGGGUGACCGGGUGGCGGCUGCGGAUGACGCAGA ...(((((((((.--------(((((((.......)))))))...))))).......(((....((((((..((.......)).(((((....)))))..)))))).))).....)))). ( -48.90) >DroSim_CAF1 106509 112 + 1 UUAUUGCUCGCUG--------UGGGGAGGAAGCCACUCCUCGAAAAGCGACUUCUUACCGAGUUGGCUGCCCGGGACUGAACCUCCUGGGUGACCGGGUGGCGCCUGCGGAUGACGCAGA .....(((((((.--------(((((((.......)))))))...)))))...............((..(((((((.......))))((....)))))..))))(((((.....))))). ( -48.00) >DroEre_CAF1 103280 120 + 1 AAAUUGCUCGCCGUAUUCAGGUGUGGAGAAAGCCACUCCCCGAAAAGCGACUUCUUACCGAGUUGGCUGCCCGGGACUGAACCUCCUGGGUGACCGGGUGGCGCCUGCGGAUGACGCAGA ...(((((((((((((((((..((((......))))..((((...((((((((......))))).)))...)))).)))))((.(((((....))))).))....))))).))).)))). ( -49.90) >DroYak_CAF1 105579 120 + 1 AAAUUGCUCGCCGUAUUCAGGUGUGGAGAAAGCCACUCCCCGAAAAGCGACUUCUUACCGAGUUAGCUGCCCGGGACUGAACCUCCUGGGUGACCGGGUGGCGCCUGCGGAUGACGCAGA ...(((((((((((((((((..((((......))))..((((...((((((((......))))).)))...)))).)))))((.(((((....))))).))....))))).))).)))). ( -49.90) >consensus UUAUUGCUCGCUGUAUUCAGGUGUGGAGAAAGCCACUCCCCGAAAAGCGACUUCUUACCGAGUUGGCUGCCCGGGACUGAACCUCCUGGGUGACCGGGUGGCGCCUGCGGAUGACGCAGA ...((.(.((((.......)))).).))...(((((((..........(((((......)))))((..((((((((.......))))))))..)))))))))..(((((.....))))). (-38.12 = -39.44 + 1.32)

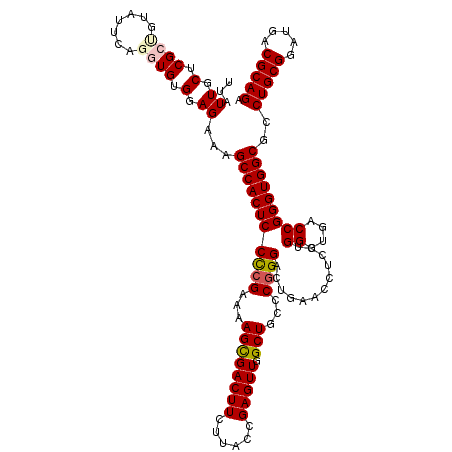

| Location | 11,367,880 – 11,368,000 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.50 |
| Mean single sequence MFE | -51.76 |
| Consensus MFE | -48.48 |
| Energy contribution | -49.24 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.765382 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11367880 120 + 27905053 CGAAAAGUGACUUCUUACCGAGUUGGCUGCCCUGGACUGAACCUCCUGGGUGACCGGGUGGCGCCUGCGGAUGACGCAGACGCACGAGCCACAGCCGCCCUCCAGGCACAUGUACUUGGU ................(((((((.(((((...(((..((...(.(((((....))))).)(((.(((((.....))))).))).))..))))))))(((.....)))......))))))) ( -49.30) >DroSec_CAF1 115510 120 + 1 CGAAAAGCGACUUCUUACCGAGUUGGCUGCACGGGACUGAACCUCCUGGGUGACCGGGUGGCGGCUGCGGAUGACGCAGACGCAGGAGCCACAGCCGCCCUCCAGGCACAUGUACUUGGU ................(((((((.(((((....((.(((..((.(((((....))))).))((.(((((.....))))).)))))...)).)))))(((.....)))......))))))) ( -49.40) >DroSim_CAF1 106541 120 + 1 CGAAAAGCGACUUCUUACCGAGUUGGCUGCCCGGGACUGAACCUCCUGGGUGACCGGGUGGCGCCUGCGGAUGACGCAGACGCAGGAGCCACAGCCGCCCUCCAGGCACAUGUACUUGGU ................(((((((.((((((((((((.......))))))))......(((((.((((((..((...))..)))))).)))))))))(((.....)))......))))))) ( -54.50) >DroEre_CAF1 103320 120 + 1 CGAAAAGCGACUUCUUACCGAGUUGGCUGCCCGGGACUGAACCUCCUGGGUGACCGGGUGGCGCCUGCGGAUGACGCAGACGCAGGAGCCGCAGCCGCCCUCCAGGCACAUGUACUUGGU ................(((((((.((((((..((.......)).(((((....))))).(((.((((((..((...))..)))))).)))))))))(((.....)))......))))))) ( -54.80) >DroYak_CAF1 105619 120 + 1 CGAAAAGCGACUUCUUACCGAGUUAGCUGCCCGGGACUGAACCUCCUGGGUGACCGGGUGGCGCCUGCGGAUGACGCAGACGCAGGAGCCGCAGCCGCCCUCCAGGCACAUGUACUUGGU ................(((((((..(((((..((.......)).(((((....))))).(((.((((((..((...))..)))))).)))))))).(((.....)))......))))))) ( -50.80) >consensus CGAAAAGCGACUUCUUACCGAGUUGGCUGCCCGGGACUGAACCUCCUGGGUGACCGGGUGGCGCCUGCGGAUGACGCAGACGCAGGAGCCACAGCCGCCCUCCAGGCACAUGUACUUGGU ................(((((((.((((((((((((.......))))))))......(((((.((((((..((...))..)))))).)))))))))(((.....)))......))))))) (-48.48 = -49.24 + 0.76)


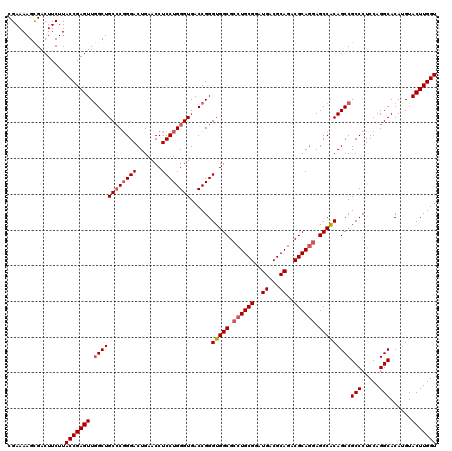
| Location | 11,367,920 – 11,368,040 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.83 |
| Mean single sequence MFE | -65.80 |
| Consensus MFE | -60.96 |
| Energy contribution | -64.36 |
| Covariance contribution | 3.40 |
| Combinations/Pair | 1.05 |
| Mean z-score | -3.10 |
| Structure conservation index | 0.93 |
| SVM decision value | 4.06 |
| SVM RNA-class probability | 0.999780 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11367920 120 + 27905053 ACCUCCUGGGUGACCGGGUGGCGCCUGCGGAUGACGCAGACGCACGAGCCACAGCCGCCCUCCAGGCACAUGUACUUGGUGGCCGUCAGGUGCAGGUGCUCCCGCAGGAAGGUGUUGAGG (((((((((....)((((.((((((((((..((((((....))..(.(((((.(((........))).((......))))))))))))..)))))))))))))))))).))))....... ( -59.50) >DroSec_CAF1 115550 120 + 1 ACCUCCUGGGUGACCGGGUGGCGGCUGCGGAUGACGCAGACGCAGGAGCCACAGCCGCCCUCCAGGCACAUGUACUUGGUGGCCGUCAGGUGCAGGUGCUCCCGGAGGAAGGCGUUGAGG .((((((((....))))(((((..(((((..((...))..)))))..))))).(((..(((((.(((((.(((((((((......))))))))).)))))...)))))..)))...)))) ( -65.90) >DroSim_CAF1 106581 120 + 1 ACCUCCUGGGUGACCGGGUGGCGCCUGCGGAUGACGCAGACGCAGGAGCCACAGCCGCCCUCCAGGCACAUGUACUUGGUGGCCGUCAGGUGCAGGUGCUCCCGGAGGAAGGCGUUGAGG .((((((((....))))(((((.((((((..((...))..)))))).))))).(((..(((((.(((((.(((((((((......))))))))).)))))...)))))..)))...)))) ( -69.30) >DroEre_CAF1 103360 120 + 1 ACCUCCUGGGUGACCGGGUGGCGCCUGCGGAUGACGCAGACGCAGGAGCCGCAGCCGCCCUCCAGGCACAUGUACUUGGUGGCCGUCAGGUGCAGGUGCUCCCGGAGAAAGGCGUUGAGG .((((((((....))))(((((.((((((..((...))..)))))).))))).(((...((((.(((((.(((((((((......))))))))).)))))...))))...)))...)))) ( -65.40) >DroYak_CAF1 105659 120 + 1 ACCUCCUGGGUGACCGGGUGGCGCCUGCGGAUGACGCAGACGCAGGAGCCGCAGCCGCCCUCCAGGCACAUGUACUUGGUGGCCGUCAGGUGCAGGUGCUCCCGGAGGAAGGCGUUGAGG .((((((((....))))(((((.((((((..((...))..)))))).))))).(((..(((((.(((((.(((((((((......))))))))).)))))...)))))..)))...)))) ( -68.90) >consensus ACCUCCUGGGUGACCGGGUGGCGCCUGCGGAUGACGCAGACGCAGGAGCCACAGCCGCCCUCCAGGCACAUGUACUUGGUGGCCGUCAGGUGCAGGUGCUCCCGGAGGAAGGCGUUGAGG .((((((((....))))(((((.((((((..((...))..)))))).))))).(((..(((((.(((((.(((((((((......))))))))).)))))...)))))..)))...)))) (-60.96 = -64.36 + 3.40)


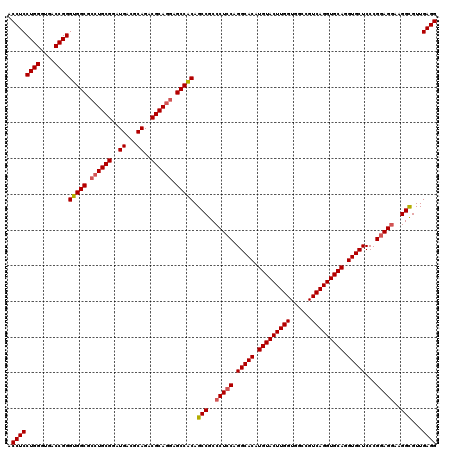
| Location | 11,367,920 – 11,368,040 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.83 |
| Mean single sequence MFE | -54.16 |
| Consensus MFE | -51.08 |
| Energy contribution | -51.32 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.94 |
| SVM decision value | 1.70 |
| SVM RNA-class probability | 0.972634 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11367920 120 - 27905053 CCUCAACACCUUCCUGCGGGAGCACCUGCACCUGACGGCCACCAAGUACAUGUGCCUGGAGGGCGGCUGUGGCUCGUGCGUCUGCGUCAUCCGCAGGCGCCACCCGGUCACCCAGGAGGU .........(((((((((((.((....)).))))..((((.(((.(((....))).))).(((.(((....((....))(((((((.....)))))))))).)))))))...))))))). ( -54.30) >DroSec_CAF1 115550 120 - 1 CCUCAACGCCUUCCUCCGGGAGCACCUGCACCUGACGGCCACCAAGUACAUGUGCCUGGAGGGCGGCUGUGGCUCCUGCGUCUGCGUCAUCCGCAGCCGCCACCCGGUCACCCAGGAGGU ............((((((((.((((.((.((.((........)).)).)).))))((((..(((((((((((.....(((....)))...)))))))))))..))))...))).))))). ( -55.10) >DroSim_CAF1 106581 120 - 1 CCUCAACGCCUUCCUCCGGGAGCACCUGCACCUGACGGCCACCAAGUACAUGUGCCUGGAGGGCGGCUGUGGCUCCUGCGUCUGCGUCAUCCGCAGGCGCCACCCGGUCACCCAGGAGGU ((((...(((((((..((((.((....)).))))..(((((.........)).))).)))))))(((((((((.((((((..((...))..)))))).)))))..))))......)))). ( -55.00) >DroEre_CAF1 103360 120 - 1 CCUCAACGCCUUUCUCCGGGAGCACCUGCACCUGACGGCCACCAAGUACAUGUGCCUGGAGGGCGGCUGCGGCUCCUGCGUCUGCGUCAUCCGCAGGCGCCACCCGGUCACCCAGGAGGU ............((((((((.((....)).......((((.(((.(((....))).))).(((.(((.((((...))))(((((((.....)))))))))).))))))).))).))))). ( -52.10) >DroYak_CAF1 105659 120 - 1 CCUCAACGCCUUCCUCCGGGAGCACCUGCACCUGACGGCCACCAAGUACAUGUGCCUGGAGGGCGGCUGCGGCUCCUGCGUCUGCGUCAUCCGCAGGCGCCACCCGGUCACCCAGGAGGU ............((((((((.((....)).......((((.(((.(((....))).))).(((.(((.((((...))))(((((((.....)))))))))).))))))).))).))))). ( -54.30) >consensus CCUCAACGCCUUCCUCCGGGAGCACCUGCACCUGACGGCCACCAAGUACAUGUGCCUGGAGGGCGGCUGUGGCUCCUGCGUCUGCGUCAUCCGCAGGCGCCACCCGGUCACCCAGGAGGU ((((...(((((((..((((.((....)).))))..(((((.........)).))).)))))))(((((.((.....(((((((((.....)))))))))..)))))))......)))). (-51.08 = -51.32 + 0.24)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:22:33 2006