
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 11,365,061 – 11,365,261 |
| Length | 200 |
| Max. P | 0.802197 |

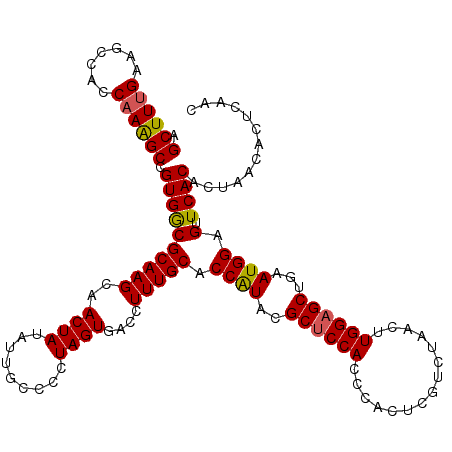
| Location | 11,365,061 – 11,365,181 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.67 |
| Mean single sequence MFE | -30.27 |
| Consensus MFE | -25.02 |
| Energy contribution | -24.94 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.779637 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11365061 120 + 27905053 AGCUUUGAAGCCACCAAAGCCGUGGCGCAAGCAACUAUAUUGCCUCUAGUGACCUUUGCACCAUACGCUCCACCCACUCGUCUAACUUGGAGCUGAAUGGAGUUCACACUAACUUUCAAC ....((((((((((.......)))))((((.........))))...(((((......((.((((.(((((((...............)))))).).)))).))...)))))...))))). ( -28.96) >DroSec_CAF1 112692 120 + 1 AGCUUUGAAGCCACCAAAGCCGUGACGCAAGCAACUAUAUUGCCCCUAGUGACCUUUGCACCAUACGCUCCACCCACUCGUCUAACUUGGAGCUGAAUGGAGUUCACACUAACACUCAAC .((((((.......)))))).(((..((((.........))))...(((((......((.((((.(((((((...............)))))).).)))).))...))))).)))..... ( -27.86) >DroSim_CAF1 103726 120 + 1 AGCUUUGAAGCCACCAAAGCCGUGGCGCAAGCAACUAUAUUGCCCCUAGUGACCUUUGCACCAUACGCUCCACCCACUCGUCUAACUUGGAGCUGAAUGGAGUUCACACUAACACUCAAC .((((((.......)))))).(((((....))..............(((((......((.((((.(((((((...............)))))).).)))).))...))))).)))..... ( -30.56) >DroEre_CAF1 100043 120 + 1 AGCUUCGAAGCAACCAAGGCCGUGGCGCAAGCCACUAAAUUGCCCCUAGUGACCUUUGCGCCGUAUGCCCCACCCACUCGUCUAACUUGGAGCUGAAUGGAGUUCACACUAACCCUCAGC .(((....)))..(((((...(((((....)))))............((((......(((.....)))......)))).......))))).(((((..((.(((......)))))))))) ( -31.60) >DroYak_CAF1 102773 120 + 1 AGCUUUGAAGCCACCAAAGCCGUGGCGCAAGCCACUAAAUUGCACCUAGUAACCUUUGCGCCAUAUGCUCCACCCACUCGUCUAACUUGGAGCUGAAUGGAGUUCACAGUGACCCCCAAC .((((((.......)))))).((((((((((..((((.........))))....))))))))))..((((((...............))))))....(((.(.((.....)).).))).. ( -32.36) >consensus AGCUUUGAAGCCACCAAAGCCGUGGCGCAAGCAACUAUAUUGCCCCUAGUGACCUUUGCACCAUACGCUCCACCCACUCGUCUAACUUGGAGCUGAAUGGAGUUCACACUAACACUCAAC .((((((.......)))))).((((((((((..((((.........))))....))))).((((..((((((...............))))))...)))).).))))............. (-25.02 = -24.94 + -0.08)

| Location | 11,365,061 – 11,365,181 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.67 |
| Mean single sequence MFE | -41.70 |
| Consensus MFE | -35.92 |
| Energy contribution | -35.24 |
| Covariance contribution | -0.68 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.802197 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11365061 120 - 27905053 GUUGAAAGUUAGUGUGAACUCCAUUCAGCUCCAAGUUAGACGAGUGGGUGGAGCGUAUGGUGCAAAGGUCACUAGAGGCAAUAUAGUUGCUUGCGCCACGGCUUUGGUGGCUUCAAAGCU .............(((..(((((((((.(((..........))))))))))))..)))((((((..(((.((((.........)))).)))))))))..((((((((.....)))))))) ( -37.00) >DroSec_CAF1 112692 120 - 1 GUUGAGUGUUAGUGUGAACUCCAUUCAGCUCCAAGUUAGACGAGUGGGUGGAGCGUAUGGUGCAAAGGUCACUAGGGGCAAUAUAGUUGCUUGCGUCACGGCUUUGGUGGCUUCAAAGCU ((((((((..(((....))).)))))))).(((.(.....)...)))(((..((...((((((....).))))).(((((((...)))))))))..)))((((((((.....)))))))) ( -36.20) >DroSim_CAF1 103726 120 - 1 GUUGAGUGUUAGUGUGAACUCCAUUCAGCUCCAAGUUAGACGAGUGGGUGGAGCGUAUGGUGCAAAGGUCACUAGGGGCAAUAUAGUUGCUUGCGCCACGGCUUUGGUGGCUUCAAAGCU ((((((((..(((....))).)))))))).(((.(.....)...)))((((.((...((((((....).))))).(((((((...))))))))).))))((((((((.....)))))))) ( -38.30) >DroEre_CAF1 100043 120 - 1 GCUGAGGGUUAGUGUGAACUCCAUUCAGCUCCAAGUUAGACGAGUGGGUGGGGCAUACGGCGCAAAGGUCACUAGGGGCAAUUUAGUGGCUUGCGCCACGGCCUUGGUUGCUUCGAAGCU ((..(((....(((((..(((((((((.(((..........)))))))))))))))))((((((..((((((((((.....))))))))))))))))....)))..)).(((....))). ( -49.90) >DroYak_CAF1 102773 120 - 1 GUUGGGGGUCACUGUGAACUCCAUUCAGCUCCAAGUUAGACGAGUGGGUGGAGCAUAUGGCGCAAAGGUUACUAGGUGCAAUUUAGUGGCUUGCGCCACGGCUUUGGUGGCUUCAAAGCU (((.((((((((((((..(((((((((.(((..........))))))))))))))).(((((((..(((((((((((...)))))))))))))))))).......)))))))))..))). ( -47.10) >consensus GUUGAGGGUUAGUGUGAACUCCAUUCAGCUCCAAGUUAGACGAGUGGGUGGAGCGUAUGGUGCAAAGGUCACUAGGGGCAAUAUAGUUGCUUGCGCCACGGCUUUGGUGGCUUCAAAGCU .............(((..(((((((((.(((..........))))))))))))..)))((((((..(((.((((.........)))).)))))))))..((((((((.....)))))))) (-35.92 = -35.24 + -0.68)

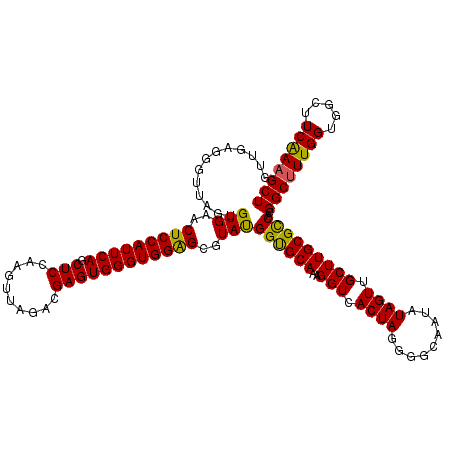

| Location | 11,365,141 – 11,365,261 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.50 |
| Mean single sequence MFE | -39.21 |
| Consensus MFE | -35.06 |
| Energy contribution | -35.42 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.13 |
| Structure conservation index | 0.89 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.510682 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11365141 120 + 27905053 UCUAACUUGGAGCUGAAUGGAGUUCACACUAACUUUCAACAUGUGGGCAAUCGCUCCUUGGGUGGCGUCCAUCCAUUGGGUGGCGCAGUAUACCUGCAGAAUGUUAUCAAGCGGCACCAC .......(((.((((..(((.(((((((.............)))))))..((((((..(((((((...)))))))..)))))).((((.....)))).........)))..)))).))). ( -38.02) >DroSec_CAF1 112772 120 + 1 UCUAACUUGGAGCUGAAUGGAGUUCACACUAACACUCAACAUGUGGGCAAUCGCUCCUUGGGUGGCGUCCAUCCAUUGGGUGGCGCAGUAUACCUGCAGAAUGUUAUCCAGCGGCACCAC .......(((.((((..(((((((((((.............)))))))..((((((..(((((((...)))))))..)))))).((((.....)))).........)))).)))).))). ( -40.42) >DroSim_CAF1 103806 120 + 1 UCUAACUUGGAGCUGAAUGGAGUUCACACUAACACUCAACAUGUGGGCAAUCGCUCCUUGGGUGGCGUCCAUCCAUUGGGUGGCGCAGUAUACCUGCAGAAUGUUAUCCAGCGGCACCAC .......(((.((((..(((((((((((.............)))))))..((((((..(((((((...)))))))..)))))).((((.....)))).........)))).)))).))). ( -40.42) >DroEre_CAF1 100123 120 + 1 UCUAACUUGGAGCUGAAUGGAGUUCACACUAACCCUCAGCAUGCGGGCGAUCGCUCCUUGGGUGGAGUUCAUCCAUUGGGUGGCACAGUAUACCUGUAGAAUGUCAUCCAACGGCACCAC .......(((.(((((..((.(((......)))))))))).(((((..((..(((((......)))))))..)).(((((((((((((.....)))).....)))))))))..)))))). ( -38.90) >DroYak_CAF1 102853 120 + 1 UCUAACUUGGAGCUGAAUGGAGUUCACAGUGACCCCCAACAUGUGGGCAAUCGCUCCCUGGGUGCAAUCCAUCCAUUGGGUGGCACAGUAUACCUGCAGGAUGUUGUCCAACGGCACCAC .......(((.((((..(((((((((((((........)).)))))))....((..((((.((((.(((((.....))))).))))((.....)).))))..))..)))).)))).))). ( -38.30) >consensus UCUAACUUGGAGCUGAAUGGAGUUCACACUAACACUCAACAUGUGGGCAAUCGCUCCUUGGGUGGCGUCCAUCCAUUGGGUGGCGCAGUAUACCUGCAGAAUGUUAUCCAGCGGCACCAC .......(((.((((..(((((((((((.............)))))))..((((((..((((((.....))))))..)))))).((((.....)))).........)))).)))).))). (-35.06 = -35.42 + 0.36)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:22:20 2006