
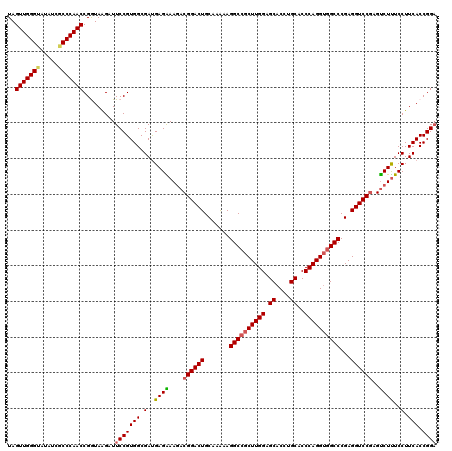
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 11,364,063 – 11,364,383 |
| Length | 320 |
| Max. P | 0.999649 |

| Location | 11,364,063 – 11,364,183 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.83 |
| Mean single sequence MFE | -49.14 |
| Consensus MFE | -45.68 |
| Energy contribution | -45.88 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.93 |
| SVM decision value | 3.83 |
| SVM RNA-class probability | 0.999649 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11364063 120 + 27905053 UAGUUGGGGAUAUCCCCCAACCGGUAAGAUUCCGUGGCGAUGAGAAACACGGACUGCAAAAAGGCCGCUUGGAGCACCUGCACCCAGGAGGCCCGAGGUCCGAGUCUUUCCUUCACCGGA ...((((((.....))))))((((((((((((.(((.(.....)...)))(((((.......((((.(((((.((....))..))))).))))...))))))))))))......))))). ( -48.60) >DroSec_CAF1 111701 120 + 1 UAGUUGGGUAUAUCGCCCAACCGGUAAGAUUCCGUGGCGAUGAGAAAGACGGACUGCAAAAAGGCCGCUUGGAGCACCUGCACCCAGGUCGCCCGAGGUCCGAGUCUUUCCUUCACCGGA ..(((((((.....)))))))(((.......)))(((.((.(.(((((((((((.((......))..(((((.(((((((....))))).)))))))))))..)))))))).)).))).. ( -47.00) >DroSim_CAF1 102735 120 + 1 UAGUUGGGUAUAUCGCCCAACCGGUAAGAUUCCGUGGCGAUGAGAAAGACGGACUGCAAAAAGGCCGCUUGGAGCACCUGCACCCAGGUGGCCCGAGGUCCAAGUCUUUCCUUCACCGGA ..(((((((.....)))))))(((.......)))(((.((.(.((((((((((((.......((((((((((.((....))..))))))))))...)))))..)))))))).)).))).. ( -52.00) >DroEre_CAF1 99050 120 + 1 UAGUUGGGUAUAUCCCCCAACCGAUAAGAUUCCGUGGCGAUGAGGAAGACGGACUGCAAAAAGGCCGCUUGGAGCACCUGCACCCAGGUGGCCCGAGGUCCGAGCCUUUCCUUCACCGGA ..((((((.......)))))).........(((((((.(..((((....((((((.......((((((((((.((....))..))))))))))...))))))..))))..).))).)))) ( -48.10) >DroYak_CAF1 101781 120 + 1 UAGUUGGGUAUAUCGCCCAACCGAUAGGAUUCCGUGGCGACGAGGAAGACGGACUGCAAGAAGGCCGCUUGGAGCACCUGCACCCAGGUGGCCCGAGGUCCGAGUCUCUCCUUCACCGGC ..(((((((.....))))))).((.((((((((.((....)).))))..((((((.......((((((((((.((....))..))))))))))...))))))......))))))...... ( -50.00) >consensus UAGUUGGGUAUAUCGCCCAACCGGUAAGAUUCCGUGGCGAUGAGAAAGACGGACUGCAAAAAGGCCGCUUGGAGCACCUGCACCCAGGUGGCCCGAGGUCCGAGUCUUUCCUUCACCGGA ..(((((((.....))))))).........(((((((.(..((((....((((((.......((((((((((.((....))..))))))))))...))))))..))))..).))).)))) (-45.68 = -45.88 + 0.20)


| Location | 11,364,063 – 11,364,183 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.83 |
| Mean single sequence MFE | -47.88 |
| Consensus MFE | -44.00 |
| Energy contribution | -44.68 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.92 |
| SVM decision value | 2.26 |
| SVM RNA-class probability | 0.991295 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11364063 120 - 27905053 UCCGGUGAAGGAAAGACUCGGACCUCGGGCCUCCUGGGUGCAGGUGCUCCAAGCGGCCUUUUUGCAGUCCGUGUUUCUCAUCGCCACGGAAUCUUACCGGUUGGGGGAUAUCCCCAACUA ...(((((..((.((((.(((((..((((((.(.((((.((....)))))).).)))))....)..))))).)))).)).))))).(((.......)))(((((((.....))))))).. ( -49.80) >DroSec_CAF1 111701 120 - 1 UCCGGUGAAGGAAAGACUCGGACCUCGGGCGACCUGGGUGCAGGUGCUCCAAGCGGCCUUUUUGCAGUCCGUCUUUCUCAUCGCCACGGAAUCUUACCGGUUGGGCGAUAUACCCAACUA ...(((((.((((((((..((((...((((.(((((....))))))).))..((((.....)))).))))))))))))..))))).(((.......)))((((((.......)))))).. ( -47.20) >DroSim_CAF1 102735 120 - 1 UCCGGUGAAGGAAAGACUUGGACCUCGGGCCACCUGGGUGCAGGUGCUCCAAGCGGCCUUUUUGCAGUCCGUCUUUCUCAUCGCCACGGAAUCUUACCGGUUGGGCGAUAUACCCAACUA ...(((((.((((((((..((((...((..((((((....))))))..))..((((.....)))).))))))))))))..))))).(((.......)))((((((.......)))))).. ( -48.60) >DroEre_CAF1 99050 120 - 1 UCCGGUGAAGGAAAGGCUCGGACCUCGGGCCACCUGGGUGCAGGUGCUCCAAGCGGCCUUUUUGCAGUCCGUCUUCCUCAUCGCCACGGAAUCUUAUCGGUUGGGGGAUAUACCCAACUA (((((((((((((.(((..((((...((..((((((....))))))..))..((((.....)))).))))))))))))..)))))..)))........(((((((.......))))))). ( -47.60) >DroYak_CAF1 101781 120 - 1 GCCGGUGAAGGAGAGACUCGGACCUCGGGCCACCUGGGUGCAGGUGCUCCAAGCGGCCUUCUUGCAGUCCGUCUUCCUCGUCGCCACGGAAUCCUAUCGGUUGGGCGAUAUACCCAACUA ...(((((..(((..(..(((((...((..((((((....))))))..))..((((.....)))).)))))..)..))).))))).............(((((((.......))))))). ( -46.20) >consensus UCCGGUGAAGGAAAGACUCGGACCUCGGGCCACCUGGGUGCAGGUGCUCCAAGCGGCCUUUUUGCAGUCCGUCUUUCUCAUCGCCACGGAAUCUUACCGGUUGGGCGAUAUACCCAACUA ((((((((.((((((((..((((..((((((.(.((((.((....)))))).).)))))....)..))))))))))))..)))))..)))........(((((((.......))))))). (-44.00 = -44.68 + 0.68)



| Location | 11,364,183 – 11,364,303 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.33 |
| Mean single sequence MFE | -43.38 |
| Consensus MFE | -41.06 |
| Energy contribution | -41.14 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.24 |
| SVM RNA-class probability | 0.935062 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11364183 120 + 27905053 GCCAGUCGCUUGUUUAGAGUGUCGCAGGCUUUUCUCACUGCCAAUCCGAUCAUCUCGCUGGUCAUCGAGUUGGCCGUCAGCAUGGAGUUGGCUCCGUUUACAGUAUUGCUGGCCUCGACU .......((((.....))))((((.((((.......((.(((((..((((.(((.....))).))))..))))).))(((((.((((....))))(....).....))))))))))))). ( -40.50) >DroSec_CAF1 111821 120 + 1 GCCAGUCGCUUGUUCAGAGUGUCGCAGGCUUUUCUCACUGCCAAUCCGAUCAUCUCGCUGGUCAUCGAGUUGGCCGUCAGCAUGGAGUUGGCUCCGUUGACAGUAUUGCUAGCCUCCACU ....(.(((((.....))))).)(.(((((......((((((((..((((.(((.....))).))))..))))).((((((..((((....)))))))))))))......))))).)... ( -41.70) >DroSim_CAF1 102855 120 + 1 GCCAGUCGCUUGUUCAGAGUGUCGCAGGCUUUUCUCACUGCCAAUCCGAUCAUCUCGCUGGUCAUCGAGUUGGCCGUCAGCAUGGAGUUGGCUCCGUUGACGGUAUUGCUAGCCUCCACU ....(.(((((.....))))).)(.(((((......((.(((((..((((.(((.....))).))))..)))))(((((((..((((....)))))))))))))......))))).)... ( -42.30) >DroEre_CAF1 99170 120 + 1 GCCAGUCGCGUGUUCAGGGUGUCGCAGGCUUUUCUCACUGCCAGUCCGAUCAUCUCGCUGGUCAUCGAGUUGGCCGUCAGCAUGGAGUUGGCUCCGCUGACAGUAUUGCUGGCCUCCACU ((((((.(((............))).((((...(((..(((((((..(.....)..))))).))..)))..))))((((((..((((....))))))))))......))))))....... ( -45.00) >DroYak_CAF1 101901 120 + 1 GCCAGUCGCUUGUUCAGAGUGUCGCAGGCUUUUCUCACUGCCAGUCCGAUCAUCUCGCUGGUCAUCGAGUUGGCCGUCAGCAUGGAGUUGGCUCCGCUGACGGUAUUGCUGGCCUCCACU (((((((((((.....)))))..((((..........))))(((..((((.(((.....))).))))..)))(((((((((..((((....)))))))))))))...))))))....... ( -47.40) >consensus GCCAGUCGCUUGUUCAGAGUGUCGCAGGCUUUUCUCACUGCCAAUCCGAUCAUCUCGCUGGUCAUCGAGUUGGCCGUCAGCAUGGAGUUGGCUCCGUUGACAGUAUUGCUGGCCUCCACU ((((((.((((((..........)))))).......((((((((..((((.(((.....))).))))..))))).((((((..((((....)))))))))))))...))))))....... (-41.06 = -41.14 + 0.08)

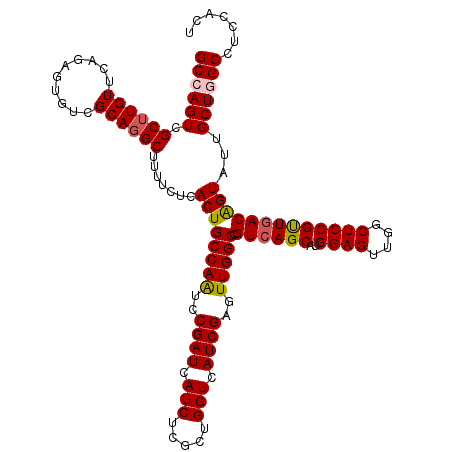

| Location | 11,364,263 – 11,364,383 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.25 |
| Mean single sequence MFE | -38.36 |
| Consensus MFE | -33.12 |
| Energy contribution | -32.72 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.827324 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11364263 120 + 27905053 CAUGGAGUUGGCUCCGUUUACAGUAUUGCUGGCCUCGACUCUCACCUGGUCCAACGGAACACCCAGCACGAAUGCUGCCACCUGGGCCGCCUUGGUGUUCACUCCUUGGCCAAUCUCAAU ..((..((((((((((((..(((.....))).....((((.......)))).))))))......(((((.((.((.(((.....))).)).)).))))).........))))))..)).. ( -36.50) >DroSec_CAF1 111901 120 + 1 CAUGGAGUUGGCUCCGUUGACAGUAUUGCUAGCCUCCACUCUCACCUGGCCCAACGGAACACCCAACACGAAAGCUGCCACCUGGGCCGCCUUGGUGUUCACUCCUUGGCCAAUCUCAAU ..((..(((((((((((((..((.....)).(((.............))).)))))))......(((((.((.((.(((.....))).)).)).))))).........))))))..)).. ( -37.72) >DroSim_CAF1 102935 120 + 1 CAUGGAGUUGGCUCCGUUGACGGUAUUGCUAGCCUCCACUCUCACCUGGUCCAACGGAACACCCAGCACGAAAGCUGCCACCUGGGCCGCCUUGGUGUUCACUCCUUGGCCAAUCUCAAU ..((((((((((.(((....)))....))))).)))))........(((.((((.(((......(((((.((.((.(((.....))).)).)).)))))...))))))))))........ ( -40.20) >DroEre_CAF1 99250 120 + 1 CAUGGAGUUGGCUCCGCUGACAGUAUUGCUGGCCUCCACUCUCACCUGAUCCAACGGAACCCCCAGCACGAAUGCUGCCACCUGGGCCGCCUUGGUGUUCACUCCUUGGCCAAUCUCUAU .((((((..(((((.(.((.((((((((((((..(((....((....))......)))....))))))...)))))).)).).)))))(((..((.(....).))..)))....)))))) ( -38.90) >DroYak_CAF1 101981 120 + 1 CAUGGAGUUGGCUCCGCUGACGGUAUUGCUGGCCUCCACUUUCACCUGAUUUAGGGGAACGCCCAGCACGAAUGCUGCCACCUGGGCCGCCUUGGUGUUCACUCCCUGGCCAAUCUCAAU ....(((..(((((.(.((.(((((((((((((.(((.((.((....))...)).)))..).))))))...)))))).)).).)))))(((..((.(....).))..)))....)))... ( -38.50) >consensus CAUGGAGUUGGCUCCGUUGACAGUAUUGCUGGCCUCCACUCUCACCUGGUCCAACGGAACACCCAGCACGAAUGCUGCCACCUGGGCCGCCUUGGUGUUCACUCCUUGGCCAAUCUCAAU ....(((((((((((((((.(((...((..((......))..)).)))...)))))))......(((((.((.((.(((.....))).)).)).))))).........))))).)))... (-33.12 = -32.72 + -0.40)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:22:17 2006